Molecular epidemiology of respiratory syncytial virus in Switzerland 2019–2024 from
nucleic acid testing and whole-genome sequencing
DOI: https://doi.org/https://doi.org/10.57187/s.4600
Alexander Kuznetsovab*,
Rainer Gosertc*,
Ulrich Heiningerd,
Nina Khannae,
Sarah Tschudin-Suttere,
Richard A. Neherab,
Karoline Leuzingerc
a Biozentrum,
University of Basel, Basel, Switzerland
b Swiss Institute of Bioinformatics, Basel,
Switzerland
c Clinical Virology, University Hospital
Basel, Basel, Switzerland
d Paediatric Infectious Diseases and
Hospital Epidemiology, University Children Hospital Basel, Basel, Switzerland
e Infectious Diseases and Hospital
Epidemiology, University Hospital Basel, Basel, Switzerland
* Equal
contribution as first authors
Summary
BACKGROUND
AND AIMS: Respiratory syncytial virus (RSV) infection
is one of the leading causes of hospitalisation in infants, the elderly and
immunocompromised patients, with significant morbidity and mortality rates.
Despite its global impact, epidemiological surveillance of RSV in Switzerland
has historically been limited compared to the USA and European countries. A
significant surge in RSV activity and hospitalisations after the COVID-19
pandemic, along with the introduction of new monoclonal antibodies and RSV
vaccines, has led to an increased interest in the molecular epidemiology of
RSV. To ensure continued efficacy of the new preventive options, monitoring of
the genetic diversity of circulating RSV strains, their evolutionary dynamics
and the potential emergence of resistance mutations is of central importance.
The present study aimed to characterise the genetic diversity and seasonal
trends of RSV dynamics in Switzerland from 2019 to 2024, mainly over the last
two post-pandemic winter seasons (2022/23 and 2023/24).
METHODS: A total of 48,897 respiratory clinical specimens from 30,782 patients
with respiratory tract infections were tested for RSV at a tertiary care centre
in Northwestern Switzerland between July 2019 and June 2024. RSV activity over
these seasons was investigated. Amplicon-based whole-genome sequencing was
performed on 182 selected samples, with 125 high-quality consensus genomes and
phylogenies reconstructed. Lineage distribution and seasonal subtype prevalence
were compared to European data.
RESULTS: RSV activity was absent during the 2020/21 pandemic season, but
surged with an off-season peak in summer 2021. RSV B predominated during the
2022/23 season, while a shift to RSV A occurred in the 2023/24 winter season,
which is in line with neighbouring European countries. Lineage subtype
distribution showed high concordance with circulating European strains. RSV A
exhibited greater diversity (mean pairwise Hamming distance 0.015, SD 0.006)
than RSV B (mean 0.006, 0.003), with all current strains falling within G protein
duplication clades A.D.1, 2, 3 and 5 for RSV A, and B.D.E.1 and B.D.4.1.1 for
RSV B.
CONCLUSION: By contributing 125 newly assembled RSV genome sequences, we have significantly
increased the number of publicly available whole-genome sequences from
Switzerland. Our study provides a genomic surveillance of RSV in Switzerland,
analysing seasonal patterns, alternating subtype dominance, and consistency
with broader European trends. A comprehensive understanding of the molecular
epidemiology of RSV enables healthcare providers and public health authorities
to monitor the effectiveness of current vaccines and monoclonal antibodies,
associate lineages and serotypes with disease severity, implement timely
interventions and detect emerging variants, ultimately reducing the burden of
RSV-related illnesses.
Introduction
Respiratory syncytial virus (RSV) is a
single-stranded RNA virus that causes respiratory tract infections. RSV
infection generally manifests with mild, cold-like symptoms but can cause
severe complications in high-risk groups such as infants, older adults and
immunocompromised patients [1].
Globally, RSV is the most common cause of lower respiratory tract infections in
infants [2].
Prior to the COVID-19 pandemic, RSV
activity in Switzerland followed a consistent seasonal pattern, typically
commencing in October, peaking in December or January and subsiding by April.
The number of hospitalisations exhibited a two-yearly regularity, where even-to-odd
winter seasons showed significantly more
hospitalisations than preceding ones [3]. However, from the first
implementation of social distancing measures in late February 2020 to the
post-pandemic winter seasons of 2022/23 and 2023/24, significant changes in RSV
epidemiology have been observed [4–6]. These changes include
shifts in the timing, intensity and age distribution of RSV infections,
reflecting the impact of public health measures and the subsequent relaxation
of non-pharmaceutical interventions. While some have hypothesised that these
shifts may be due to increased virulence of circulating RSV strains during the
pandemic [7],
current molecular epidemiological surveillance shows that post-pandemic
circulation is dominated by several lineages with pre-pandemic roots,
suggesting epidemiological rather than virological reasons [8].
With the recent introduction of monoclonal
antibodies and prefusion F protein-based vaccines (RSVpreF) in 2023/24, such as
nirsevimab (Beyfortus®) [9, 10]
by AstraZeneca and Sanofi, Abrysvo® by Pfizer [11] and Arexvy® by
GlaxoSmithKline [12],
options for specific prevention of RSV infections have increased dramatically.
These interventions raise important concerns about the molecular adaptation of
RSV under selective pressure, possibly leading to variants that escape
vaccine-induced immunity or develop resistance to monoclonal antibodies. The
main focus of attention lies on the Fusion (F) protein, the primary antigen in
current recombinant vaccines and a target for monoclonal antibodies [13]. Several
studies so
far have shown little to no elevated levels of polymorphisms in the vaccinees
compared to the control group [14]. However, RSV is a
rapidly evolving virus with a high mutation rate, and continuous monitoring is
crucial to detect potential early signals of adaptation and escape variants
that may arise as vaccines and monoclonal antibodies are rolled out more
widely. A comprehensive understanding of the molecular epidemiology of RSV
enables healthcare providers and public health authorities to monitor the
effectiveness of current vaccines and monoclonal antibodies, assess the
severity of RSV-related diseases, implement timely interventions and detect
emerging variants, ultimately reducing the burden of RSV-related illnesses.
RSV is divided into two major antigenic
subtypes, RSV A and RSV B, with the greatest divergence in the gene coding for
the attachment glycoprotein G (G protein). The estimated common ancestor of the
RSV A and RSV B circulated around 250 years ago [15]. A recent
hierarchical classification by Goya et al. [16] defined 24 and 16
lineages within RSV A and RSV B, respectively, based on phylogeny and amino
acid markers. This novel unified nomenclature is designed to be kept up-to-date
through designation of new lineages with the aim of tracking epidemiologically
relevant viral variants and comparing the circulation of the virus from season
to season and across geographies.
In this study, we present data on RSV
dynamics in Northwestern Switzerland during the pre-pandemic season 2019/20,
the pandemic seasons 2020/21 and 2021/22, and the post-pandemic periods 2022/23
and 2023/24. Throughout this timeframe, we conducted nucleic acid testing and
full-genome sequencing of RSV from symptomatic patients presenting with respiratory
tract infections at our tertiary care hospital.
Methods
Patient cohorts and inclusion criteria
Patients included in our study presented
with acute respiratory tract infections, as defined by at least 1 respiratory
and 1 systemic symptom/sign such as clogged or runny nasal airways, sore
throat, cough, fatigue, fever, headache, chills or myalgia [17]. This clinical
diagnosis is a prerequisite for ordering broad molecular multiplex panel
testing at our tertiary care hospital. The patients presented to the outpatient
or emergency departments of University Hospital Basel or University Children’s
Hospital Basel between July 2019 and June 2024 and underwent RSV-specific
nucleic acid testing. RSV-specific testing was performed using the Biofire FilmArray
RPP (bioMérieux, Marcy-l’Étoile, France) and additionally the Xpert®
Xpress-CoV-2/Flu/RSV-Plus system (Cepheid, CA, USA) [18, 19]. The sensitivity
of the Biofire FilmArray RPP for detecting RSV ranges from 95% to 100%, while
specificity exceeds 99%. The corresponding values for the Xpert®
Xpress-CoV-2/Flu/RSV-Plus system are 95–98% sensitivity and >98% specificity [20,
21].
Amplicon-based next-generation sequencing approach
The 500bp-NAT (nucleic acid testing) primers
were designed to match conserved sequences across all publicly available
full-length RSV genome sequences from the NCBI nucleotide database (totalling 3010
as of 1 January 2022, with 2002 RSV A and 1008 RSV B). NAT primers were
designed using the publicly available PRIMAL tool [22] that allows the
design of highly multiplex primer pools. Additionally, 2000bp-NAT primers and
1000bp-RSV A and RSV B subgroup-specific NAT primers were employed from [23], and
used in 2
separate primer pools. All NATs used the Iproof High Fidelity DNA Polymerase kit
(BioRad, CA, USA) with 600 nM end concentration of the different primer pools (appendix
figure S4, supplementary
files 1 and 2). NATs had a reaction volume of 25 µl containing
5 µl of extracted DNA and were run on Veriti™ Thermal Cyclers (Applied
Biosystems, MA, USA) using the thermal cycling protocol specified in the Iproof
High Fidelity DNA Polymerase kit. Library preparation was done by pooling the
six amplicons from the 2000bp, 1000bp and 500bp NAT reactions using the KAPA
HyperPrep Kit (Roche, Rotkreuz, Switzerland) following the manufacturers’
instructions. Next-generation sequencing (NGS) was performed on a MiniSeq
platform (Illumina, CA, USA). Raw data, including FASTQ files, were
subsequently processed and organised for downstream analysis.
Genome assembly
Raw sequencing reads were trimmed using
Trim Galore v0.6.10 and mapped to reference genomes with BWA-MEM v0.7.18.
Pileup analysis and consensus sequence construction were performed using custom
scripts adapted from the Enterovirus D68 project [24, 25].
Preprocessing and sample selection
A total of 182 samples were initially
selected for the analysis: 159 from outpatients and 23 from hospitalised
patients with the best coverage (appendix figure S3). Preprocessing included masking
positions with
less than 90% main allele frequency. After applying a coverage threshold of at
least 100× over 80% of the genome, 125 samples (96 RSV A, 29 RSV B) were
retained for the phylogenetic analysis.
Phylogenetic analysis
Phylogenetic trees were constructed using
the Nextstrain [26]
CLI pipeline v8.5.3, which includes Augur v26.0.0 and Auspice v2.58.0.
Consensus sequences were aligned with background data from the NCBI Virus
database using Nextclade [27]
v3.8.2, and trees were inferred using IQ-TREE [28] v2.3.6. Two trees
were generated: one with no regional or temporal filters for global context and
another focused on recent (last 3 years) European sequences for more detailed
comparison. Diversity was calculated as follows: for each RSV subtype, aligned
consensus sequences were taken; Hamming distance (the number of positions with
different nucleotides for two aligned genomes) for each pair was calculated,
excluding uncertain consensus positions and normalising to the length of the
compared part. All the tools mentioned above are publicly available under
GPL-3.0 or MIT licences.
Ethics statement
The study was conducted according to good
laboratory practice and in accordance with the Declaration of Helsinki and
national and institutional standards, and was approved by the ethics committee
of Northwestern and Central Switzerland (EKNZ 2024-00813).
Results
Pre- and post-pandemic seasonal dynamics of RSV
Patients presenting with symptoms of respiratory
tract infections at the outpatient or emergency departments of University
Hospital Basel or University Children’s Hospital Basel underwent RSV-specific
nucleic acid testing between July 2019 and June 2024 (appendix table S2). RSV-positive
samples with
sufficient residual material further underwent full-genome sequencing,
representing a convenience sample over the whole study period.
A total of 48,897 respiratory clinical
specimens from 30,782 patients with respiratory tract infections – of whom 14,613
(47.5%) were female and 6436 (20.9%) paediatric patients <18 years – were
submitted for routine RSV-specific nucleic acid testing between July 2019 and
June 2024 at our tertiary care hospital in Northwestern Switzerland (table 1, appendix
table S1). The median patient age was 62 years (range: 1 to 106
years).
Table 1Distribution of tested and sequenced samples.
| Demographic |
2019 |
2020 |
2021 |
2022 |
2023 |
2024 |
Total |
| <18 |
≥18 |
<18 |
≥18 |
<18 |
≥18 |
<18 |
≥18 |
<18 |
≥18 |
<18 |
≥18 |
<18 |
≥18 |
| RSV-positive samples (n) |
43 |
21 |
28 |
43 |
310 |
55 |
481 |
310 |
231 |
123 |
131 |
118 |
1224 |
670 |
| RSV-positive patients (n) |
43 |
21 |
18 |
30 |
252 |
55 |
433 |
248 |
214 |
98 |
117 |
99 |
1077 |
551 |
| RSV sequences (n) |
|
|
|
2 |
|
7 |
|
23 |
69 |
41 |
16 |
24 |
85 |
97 |
| RSV sequences, passed QC (n) |
|
|
|
1 |
|
3 |
|
13 |
56 |
30 |
6 |
16 |
62 |
63 |
First, we assessed the seasonality and peak
activity of RSV during the study period. In 2019, prior to the COVID-19
pandemic, the number of RSV infections in our catchment area increased in
October and subsided by February 2020, consistent with seasonal patterns
observed before the pandemic [3]. The implementation of non-pharmaceutical
interventions to reduce the spread of SARS-CoV-2 in 2020 and 2021, including
travel restrictions, social distancing, mask mandates, and school and business
closures, disrupted this regularity. Notably, during the 2020/21 season, RSV
activity was minimal, with no significant epidemic observed. However, an
atypical early surge occurred in 2021. Cases began to rise in May, peaking in
July, and persisting until January 2022. Subsequent post-pandemic 2022/23 and
2023/24 seasons indicated a return to pre-pandemic seasonality, with activity
commencing in late summer, peaking in November and declining by January (figure
1A).
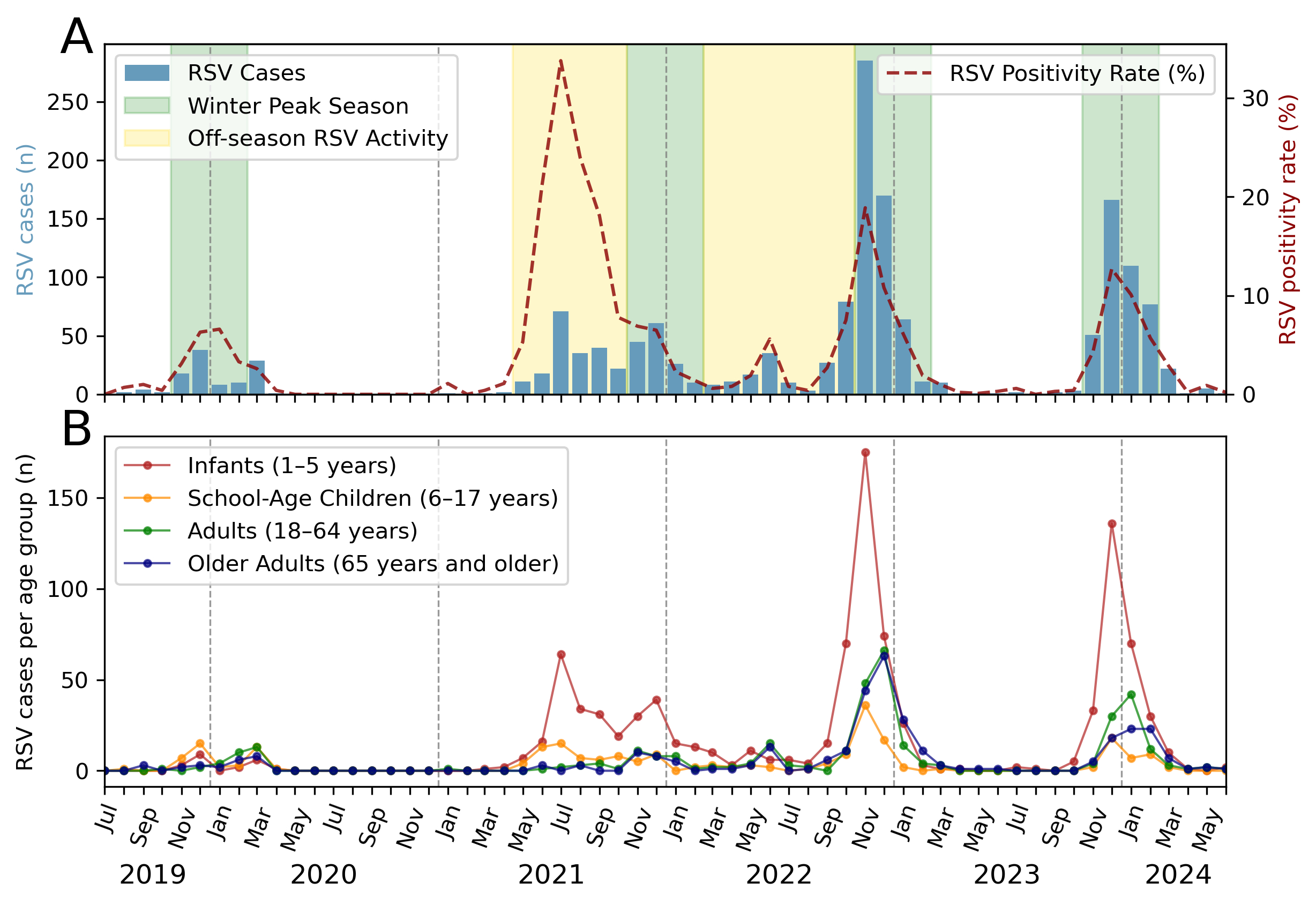
Figure 1Respiratory syncytial virus (RSV) seasonality, peak activity and
demographic distribution. (A) RSV detections during the pandemic seasons
(2019/20, 2020/21, 2021/22) and the post-pandemic period (2022/23, 2023/24).
The green background bar represents the typical RSV winter season, running from
November to February, and the yellow background bar indicates off-season RSV
activity. The number of positive cases per month is indicated by blue bars and
the positive rate is shown as a dashed red line (right axis). (B) Age
distribution of RSV cases by month for the specified period.
The off-season activity in 2021
significantly impacted paediatric patients aged 1 to 5 years, who accounted for
most RSV cases during that period. In subsequent seasons, we observed an
increase in diagnosed RSV infections among adults and older patients aged ≥65
years (figure 1B). In the 2022/23 RSV season, RSV infections in infants and
school-aged children peaked 1–2 months earlier than those in adults, which
suggests that younger populations may have had higher susceptibility in the
first post-pandemic season.
During the 2022/23 and 2023/24 seasons,
adult populations experienced consecutive major RSV epidemics, leading to
significantly increased RSV-related hospitalisations compared to the pandemic
2019/20 and 2020/21 seasons (appendix figure S1). The elevated case numbers in post-pandemic
seasons relative to pre-pandemic seasons can be attributed, in part, to
increased testing rates, as test positivity does not differ as dramatically
between pre- and post-pandemic seasons. Hospitalisation rates for paediatric
patients were not available for this study, but recent data suggest that
immunologically naive children or those with limited prior exposure to RSV
during the COVID-19 pandemic experienced more severe RSV-related illness and faced
a higher risk of hospitalisation compared to pre-pandemic years [5, 29, 30].
Seasonal distribution of RSV subtypes
Respiratory viruses generally show a
similar distribution of genotypes in geographically well-connected regions, but
during the pandemic period with travel restrictions and variable approaches to
social distancing and non-pharmaceutical interventions, resolving the local
diversity of viruses is of particular interest. Furthermore, addressing the
hypotheses that circulating RSV strains after the pandemic may have exhibited
increased virulence (appendix table S3),
potentially contributing to higher transmission rates, increased case numbers
and related hospitalisations, requires whole-genome sequencing of a
sufficiently large number of viral samples to detect or rule out strong
associations between viral genotypes and presentation.
While only a few samples were available for
whole-genome sequencing prior to 2022, we generated 20 full genomes from the
2022/23 season and 100 for the 2023/24 season. As shown in figure 2, RSV B dominated
during the 2022/23 season (19 of 20
samples), while RSV A was the main subtype in the 2023/24 season (92 of 100
samples).
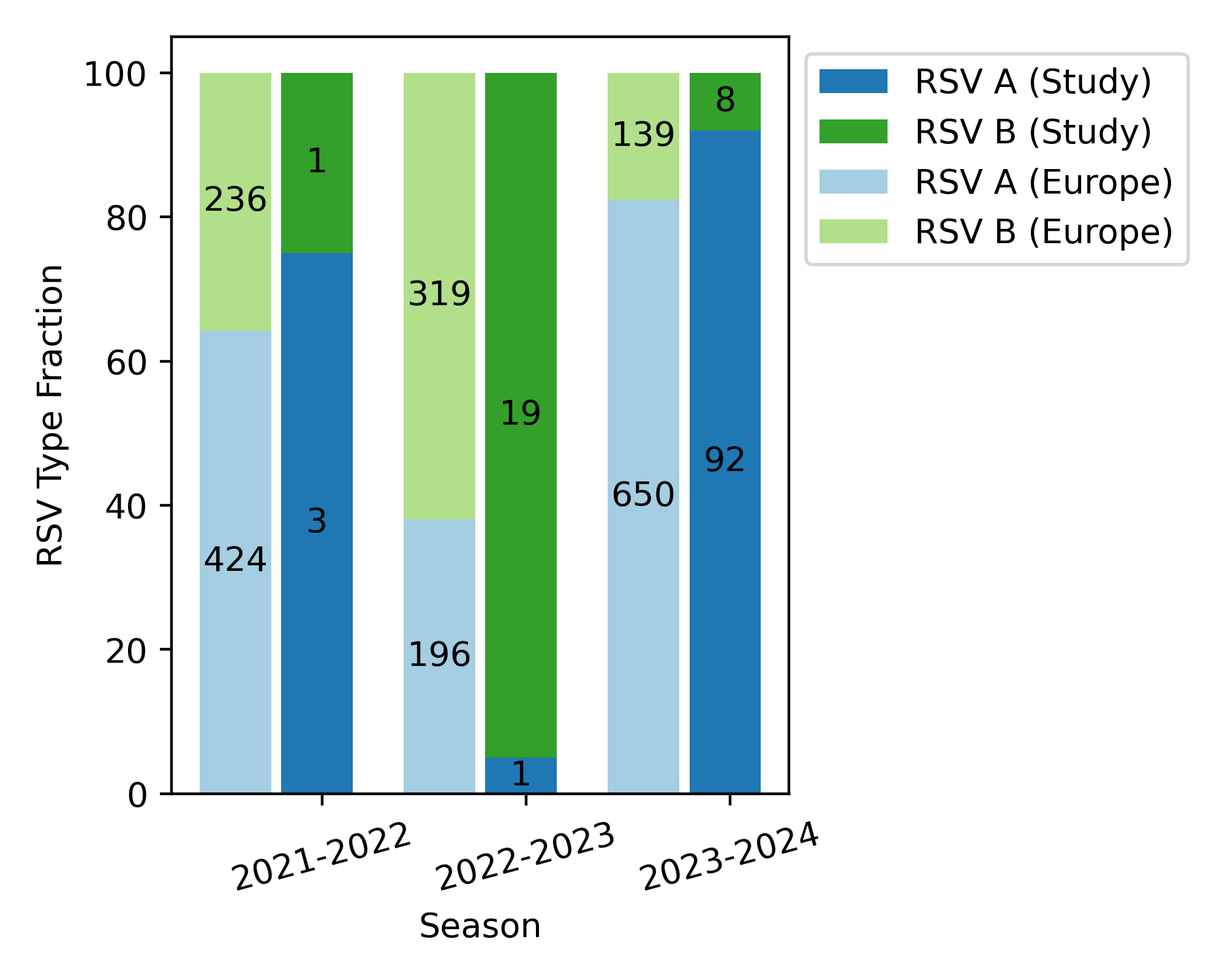
Figure 2Annual distribution of RSV A and RSV B subtypes in Switzerland and Europe
over the 2021/22, 2022/23 and 2023/24 seasons. Numbers inside the bars
display actual numbers of respiratory
syncytial virus (RSV) subtype cases, while heights correspond to
fractions. Stacked bars to the left show same-season RSV subtype distributions
in European samples available in the NCBI virus database.
Phylogenetic analysis of RSV subtypes
All currently circulating RSV strains
belong to clades with duplications in the G protein (figures 3A and 4A), which
occurred independently in RSV A and RSV B [31, 32] in 2009 and 1996,
respectively, according to the reconstructed phylogenies. These clades are
named A.D and B.D in the new lineage nomenclature [16] and have been
divided into several sublineages.
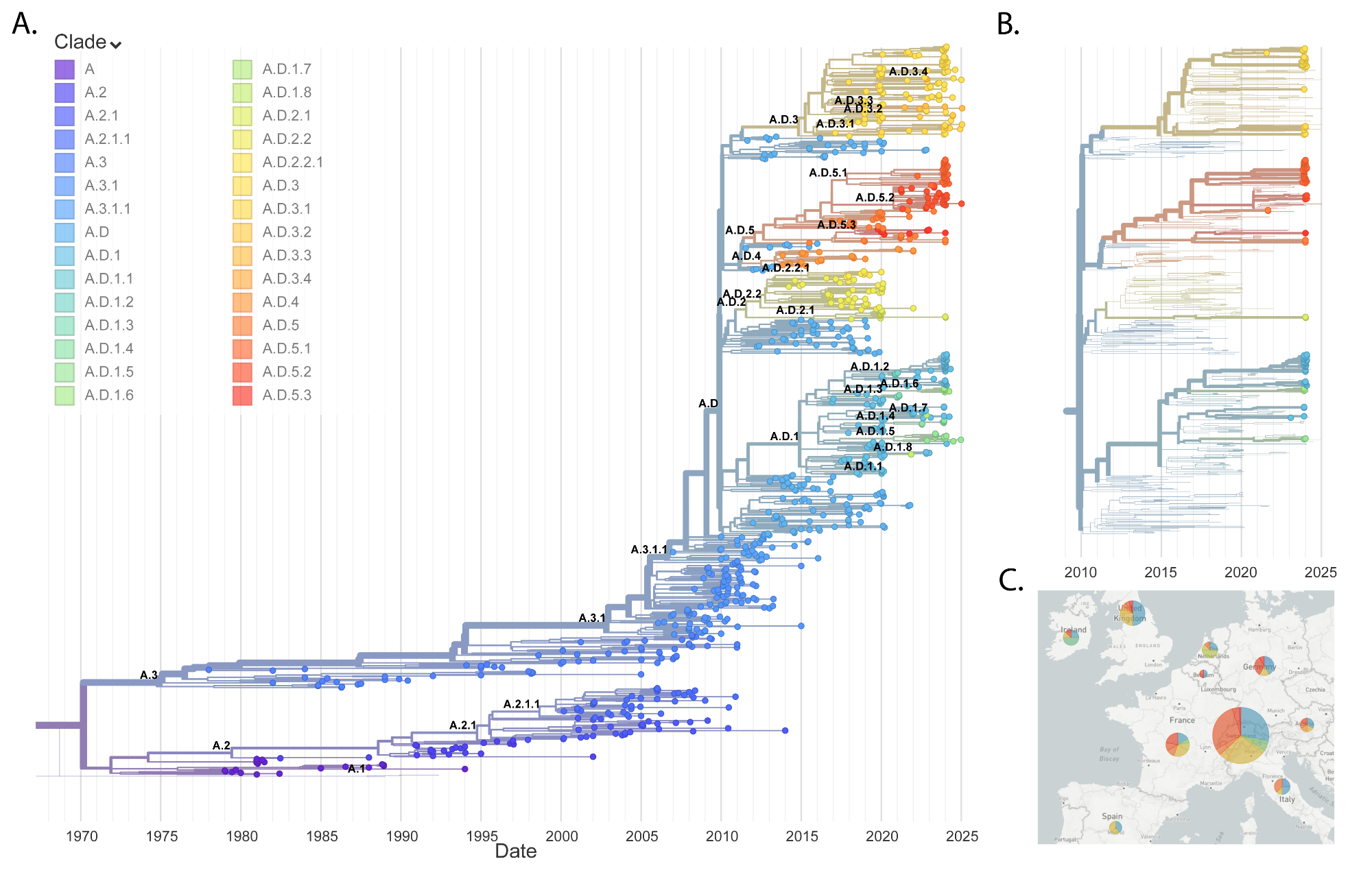
Figure 3RSV A phylogeny. (A) Global
phylogenetic tree of RSV A sequences. (B) RSV A phylogenetic tree focusing on
the last two years of European data. Bold leaves indicate sequences generated
in this study. (C) Geographic distribution of RSV A clades in Europe over the
last two seasons.
RSV A: Switzerland vs Europe
Phylogenetic analysis of RSV A sequences
revealed a high degree of correspondence between Switzerland and the broader
European context (figures 3B–C). Sequence diversity levels are higher for RSV
A, with mean pairwise Hamming distance of 0.015 and SD 0.006, compared to the
mean of 0.006 and SD 0.003 for RSV B (appendix figure S2). RSV A samples were
found in three main lineages – A.D.1, A.D.3 and A.D.5 – and, to a lesser extent,
in A.D.2. These lineages have been observed across Europe and have a common ancestor
prior to 2015. Therefore, multiple lineages of RSV A have persisted from the
pre-pandemic period and there is no indication that a particular lineage with
distinct properties has emerged that might be associated with a different
presentation. Overall, this diverse RSV A population with high correspondence
to European dynamics suggests that RSV A was introduced into Switzerland
numerous times each season and that there is frequent exchange with neighbouring
countries.
RSV B: Switzerland vs Europe
The RSV B phylogeny also showed high
correspondence with other European countries. Current circulating strains of
RSV B mostly fall within B.D.E.1 and B.D.4.1.1 clades, which is also the case
for our data (figures 4B-C, appendix table S4): 26 samples belong to B.D.E.1 and 3
samples to its
parent clade, B.D.4.1.1. The lineage B.D.E.1 emerged shortly before the
pandemic and has expanded globally over the past 5 years. This recent expansion
explains the lower Hamming distance levels compared to RSV A and the different
clade distribution. Apart from these differences between subtypes, RSV B
samples also display multiple introduction events with many lineages persisting
across many seasons.
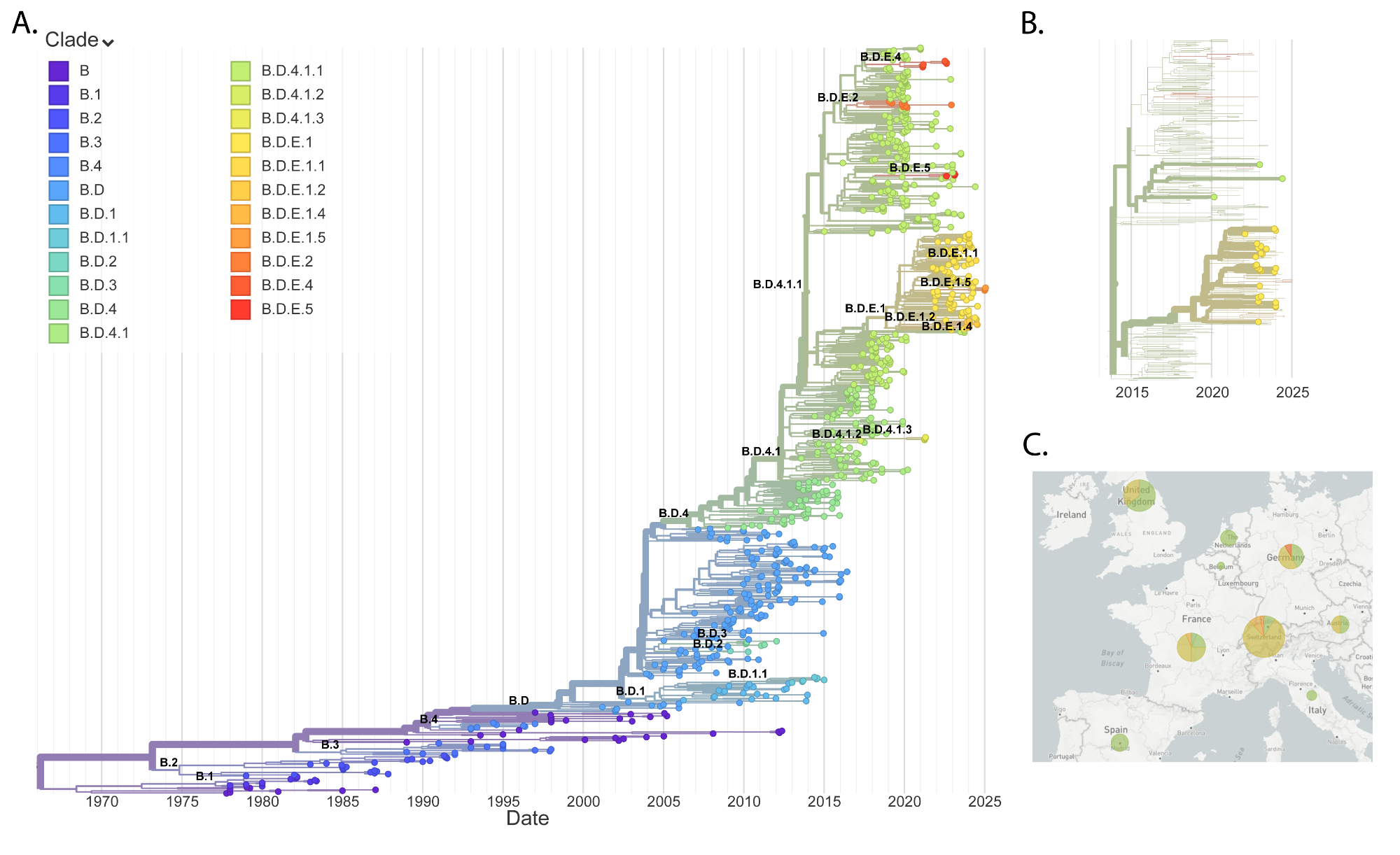
Figure 4RSV B phylogeny. (A) Global
phylogenetic tree of RSV B sequences. (B) RSV B phylogenetic tree focusing on
the last two years of European data. Bold leaves indicate sequences generated
in this study. (C) Geographic distribution of RSV B clades in Europe during the
last two seasons.
Discussion
The COVID-19 pandemic has significantly
disrupted the typical seasonal patterns of RSV transmission and activity [33]. Public
health
measures implemented to curb the spread of SARS-CoV-2, such as social
distancing, mask-wearing and lockdowns, led to a temporary decline in RSV cases [34].
Here, we reported
on the patterns of RSV infections and hospitalisations during this period in
Northwestern Switzerland. As these measures were relaxed, a notable resurgence
of RSV activity was observed. For example, an unusual outbreak occurred in the
summer of 2021, deviating from the typical winter peak. This was followed by a
strong resurgence of RSV cases and related hospitalisations in the following
2022/23 and 2023/24 seasons after all COVID-19 restrictions were lifted. The
observed patterns of RSV infections generally mirror those observed elsewhere
in Switzerland [5, 6],
but are notably different from those of the neighbouring countries Germany [35–37],
Italy [38–40] and Austria [41], where no summer
wave was observed in 2021. The USA, on the other hand, did observe significant
RSV activity between April 2021 and February 2022 [42]. Considering the
varied resurgence patterns of RSV across Europe, one might anticipate that the
dominant viral type (RSV A vs B) differs between countries, or that different
sublineages of RSV A or B circulated in different regions. To investigate this
hypothesis, we performed whole-genome sequencing on respiratory swabs,
generating a total of 125 RSV genomes. Overall, the dominance patterns of RSV A
and B were consistent with those of neighbouring countries. However, Europe as
a whole demonstrated a moderately more even distribution of both viral types,
which might be due to the much larger and diverse demographic.
Aggregated data from Europe for these two
seasons reveal the presence of the same predominant subtypes. However, the
dominance is less pronounced: RSV B accounted for 62% of the samples in the
2022/23 season (319 of 515) while RSV A comprised 82% of the samples in the
2023/24 season (650 of 789). Data from the neighbouring countries Austria and
Germany indicate a similarly pronounced alternation in subtype dominance,
exhibiting a 90% difference between these seasons [37, 41], akin to the
trends observed in Switzerland. An analysis of wastewater samples in
Switzerland found the same subtype distribution for post-pandemic seasons [43]. In
contrast, the
USA exhibits the opposite pattern in seasonal subtype dynamics: RSV B was
predominant in the 2021/22 and 2023/24 seasons, while RSV A prevailed during
the 2022/23 season [42, 44].
This disparity underscores the necessity for global surveillance of RSV.
Concordance was similarly observed at the
lineage level. For RSV A, these sequenced strains reflect lineages that were
prevalent in Europe during the same period. Most of these lineages have been
circulating in parallel since 2010, predating the pandemic, which implies that
no specific lineage was responsible for the resurgence of cases. Instead,
observed differences in disease presentation and incidence are likely
attributable to epidemiological or immunological factors arising from non-pharmaceutical
interventions and reduced exposure to viral pathogens during periods of social
distancing. Conversely, since the pandemic’s onset, RSV B diversity has
primarily been driven by lineage B.D.E.1 and its descendants, which likely
arose more recently in 2019 [45].
Consequently, RSV B sequences exhibit
greater similarity to one another compared to RSV A sequences. Notably, the
diversity that we observed in Northwestern Switzerland aligns closely with the
circulating strains in neighbouring European countries.
These findings indicate a sufficiently
rapid exchange of RSV among European countries, facilitating alignment of the
circulation patterns, even amid restricted travel and movement. However, on a
broader geographic scale, these patterns exhibit notable differences. For
instance, in the United States, different dominance patterns have been observed
for RSV A and B, reflecting a divergence in the epidemiological behaviour of
the virus compared to Europe [42].
Monitoring the circulating strains of RSV
is crucial, as RSV subtypes A and B co-circulate globally, with shifts in
predominance observed over time. Research has demonstrated that these subtypes
may present different clinical manifestations and disease outcomes. In paediatric
populations, it has been shown that RSV A infections tend to result in more
severe clinical presentations and poorer patient outcomes compared to RSV B [46–48].
Although fewer
studies have focused on adult populations, existing evidence suggests that RSV
A is associated with increased virulence, as indicated by clinical severity
scores [49].
These findings align with our observations, as the RSV A-dominant 2023/24
season coincided with an increase in hospitalisation rates, although causality
cannot be established within the scope of our study. As discussed above, the
genetic diversity of RSV A observed during the post-pandemic resurgence does
not imply that lineages that persisted through the pandemic possessed unique
characteristics.
This highlights the importance of
accurately identifying circulating RSV strains. Understanding which strains are
prevalent allows healthcare providers and public health authorities to better
anticipate potential increases in transmission rates, case numbers and disease
severity, facilitating more effective allocation of healthcare resources and
implementation of targeted interventions.
With the introduction of RSV vaccines for
adults in Switzerland, such as Arexvy and Abryso, alongside monoclonal
antibodies for children, including Beyfortus, monitoring the molecular
epidemiology of RSV and tracking escape or resistance mutations is becoming
increasingly important. While little resistance has been observed in clinical
studies to date, the widespread deployment of these preventive measures may
exert additional pressure on the virus to evolve [50]. Early
identification of such strains is essential for adapting diagnostic tools,
refining prevention strategies and enhancing vaccine strategies to maintain their
effectiveness against RSV infections.
This study has several limitations that
should be acknowledged. The limited catchment area restricts the generalisability
of the findings to broader populations, potentially overlooking regional
variations in RSV circulation. The use of convenience sampling for full genome
analysis may introduce selection bias, as samples are not randomly chosen and
may not accurately represent the entire circulating viral diversity. Additionally,
the coverage bias inherent in next-generation sequencing (NGS) results, which
excludes low-coverage samples from analysis, could influence the observed
lineage distribution by underrepresenting certain variants that are present at
lower abundance. Relying solely on consensus sequences further limits the
detection of intra-host viral diversity and minority variants, potentially
overlooking minor lineages or mutations that could impact viral evolution and
epidemiology. Lastly, detailed clinical information, including patient
outcomes, precise anatomical location of infection (upper and lower respiratory
tract infections) and disease severity, was not available in the scope of this
study. Therefore correlations between viral genotypes and these factors could
not be assessed, limiting epidemiological conclusions to subtype occurrence
analysis only. These methodological constraints may collectively influence the
study’s conclusions regarding RSV lineage dynamics and mask the full spectrum
of viral diversity within the studied population.
In summary, our analysis of RSV
epidemiology of recent seasons in Switzerland outlines the dynamics of case
numbers and subtype prevalence and highlights the importance of sustained
surveillance. Ongoing monitoring is critical to inform public health policies,
guide vaccination strategies and detect the emergence of variants with
potential clinical and epidemiological significance.
Data sharing statement
The consensus genomes generated in this
study have been submitted to the Pathoplexus database under sequence sets
PP_SS_201.2 [51] and PP_SS_202.2 [52] and to NCBI Datasets Virus Data under
Bioprojects PRJEB88486 and PRJEB88624, respectively.
Code availability: The custom algorithms
and scripts developed for this study are openly accessible and can be found in
our GitHub repository at: https://github.com/neherlab/rsv_epidemiology_2025
We encourage researchers and practitioners
to explore and use this resource for further advancements in the field.
Acknowledgments
We thank the biomedical technicians of the
Clinical Virology Department, Laboratory Medicine, University Hospital Basel,
Basel, Switzerland, for expert help and assistance. We also thank Anna Parker
for her support with data submission to the Pathoplexus genomic database.
Author contributions: RG, RAN and KL
contributed to the conceptualisation and study design. Data collection and
clinical contributions were provided by UH, NK and STS. Methodology and
laboratory analyses were conducted by RG and KL. Data analysis and
interpretation were performed by AK, RG, RAN and KL. The original draft of the
manuscript was written by AK and RAN. All authors reviewed and approved the
final manuscript.
PD Dr Karoline Leuzinger
Clinical Virology
University Hospital Basel
Petersgraben 4
CH-4031 Basel
karoline.leuzinger[at]usb.ch
References
1. Kaler J, Hussain A, Patel K, Hernandez T, Ray S. Respiratory syncytial virus: A comprehensive
review of transmission, pathophysiology, and manifestation. Cureus. 2023 Mar;15(3):e36342.
doi: https://doi.org/10.7759/cureus.36342
2. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional
mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic
analysis for the Global Burden of Disease Study 2010. Lancet. 2012 Dec;380(9859):2095–128.
doi: https://doi.org/10.1016/S0140-6736(12)61728-0
3. Stucki M, Lenzin G, Agyeman PK, Posfay-Barbe KM, Ritz N, Trück J, et al. Inpatient
burden of respiratory syncytial virus (RSV) in Switzerland, 2003 to 2021: an analysis
of administrative data. Euro Surveill. 2024 Sep;29(39):2400119. doi: https://doi.org/10.2807/1560-7917.ES.2024.29.39.2400119
4. Meslé MM, Sinnathamby M, Mook P, Pebody R, Lakhani A, Zambon M, et al.; WHO European
Region Respiratory Network Group. Seasonal and inter-seasonal RSV activity in the
European Region during the COVID-19 pandemic from autumn 2020 to summer 2022. Influenza
Other Respir Viruses. 2023 Nov;17(11):e13219. doi: https://doi.org/10.1111/irv.13219
5. Fischli K, Schöbi N, Duppenthaler A, Casaulta C, Riedel T, Kopp MV, et al. Postpandemic
fluctuations of regional respiratory syncytial virus hospitalization epidemiology:
potential impact on an immunization program in Switzerland. Eur J Pediatr. 2024 Dec;183(12):5149–61.
doi: https://doi.org/10.1007/s00431-024-05785-z
6. Sauteur PM, Plebani M, Trück J, Wagner N, Agyeman PK. Ongoing disruption of RSV epidemiology
in children in Switzerland. Lancet Reg Heal; 2024. p. 45.
7. Rao S, Armistead I, Messacar K, Alden NB, Schmoll E, Austin E, et al. Shifting epidemiology
and severity of respiratory syncytial virus in children during the COVID-19 pandemic.
JAMA Pediatr. 2023 Jul;177(7):730–2. doi: https://doi.org/10.1001/jamapediatrics.2023.1088
8. Abu-Raya B, Viñeta Paramo M, Reicherz F, Lavoie PM. Why has the epidemiology of RSV
changed during the COVID-19 pandemic? EClinicalMedicine. 2023 Jul;61:102089. doi: https://doi.org/10.1016/j.eclinm.2023.102089
9. Hammitt LL, Dagan R, Yuan Y, Baca Cots M, Bosheva M, Madhi SA, et al.; MELODY Study
Group. Nirsevimab for prevention of RSV in healthy late-preterm and term infants.
N Engl J Med. 2022 Mar;386(9):837–46. doi: https://doi.org/10.1056/NEJMoa2110275
10. Griffin MP, Yuan Y, Takas T, Domachowske JB, Madhi SA, Manzoni P, et al.; Nirsevimab
Study Group. Single-dose nirsevimab for prevention of RSV in preterm infants. N Engl
J Med. 2020 Jul;383(5):415–25. doi: https://doi.org/10.1056/NEJMoa1913556
11. Simões EA, Pahud BA, Madhi SA, Kampmann B, Shittu E, Radley D, et al.; MATISSE (Maternal
Immunization Study for Safety and Efficacy) Clinical Trial Group. Efficacy, Safety,
and Immunogenicity of the MATISSE (Maternal Immunization Study for Safety and Efficacy)
Maternal Respiratory Syncytial Virus Prefusion F Protein Vaccine Trial. Obstet Gynecol.
2025 Feb;145(2):157–67. doi: https://doi.org/10.1097/AOG.0000000000005816
12. Shirley M. RSVPreF3 OA respiratory syncytial virus vaccine in older adults: a profile
of its use. Drugs Ther Perspect. 2025;41(1):1–8. doi: https://doi.org/10.1007/s40267-024-01125-1
13. Papi A, Ison MG, Langley JM, Lee DG, Leroux-Roels I, Martinon-Torres F, et al.; AReSVi-006
Study Group. Respiratory syncytial virus prefusion F protein vaccine in older adults.
N Engl J Med. 2023 Feb;388(7):595–608. doi: https://doi.org/10.1056/NEJMoa2209604
14. Fourati S, Reslan A, Bourret J, Casalegno JS, Rahou Y, Chollet L, et al. Genotypic
and phenotypic characterisation of respiratory syncytial virus after nirsevimab breakthrough
infections: a large, multicentre, observational, real-world study. Lancet Infect Dis.
2024.
15. Saito M, Tsukagoshi H, Sada M, Sunagawa S, Shirai T, Okayama K, et al. Detailed evolutionary
analyses of the F gene in the respiratory syncytial virus subgroup A. Viruses. 2021 Dec;13(12):2525.
doi: https://doi.org/10.3390/v13122525
16. Goya S, Ruis C, Neher RA, Meijer A, Aziz A, Hinrichs AS, et al. Standardized phylogenetic
classification of human respiratory syncytial virus below the subgroup level. Emerg
Infect Dis. 2024 Aug;30(8):1631–41. doi: https://doi.org/10.3201/eid3008.240209
17. Ison MG, Hirsch HH. Community-acquired respiratory viruses in transplant patients:
diversity, impact, unmet clinical needs. Clin Microbiol Rev. 2019 Sep;32(4):10–1128.
doi: https://doi.org/10.1128/CMR.00042-19
18. Goldenberger D, Leuzinger K, Sogaard KK, Gosert R, Roloff T, Naegele K, et al. Brief
validation of the novel GeneXpert Xpress SARS-CoV-2 PCR assay. J Virol Methods. 2020 Oct;284:113925.
doi: https://doi.org/10.1016/j.jviromet.2020.113925
19. Leuzinger K, Roloff T, Gosert R, Sogaard K, Naegele K, Rentsch K, et al. Epidemiology
of severe acute respiratory syndrome coronavirus 2 emergence amidst community-acquired
respiratory viruses. J Infect Dis. 2020 Sep;222(8):1270–9. doi: https://doi.org/10.1093/infdis/jiaa464
20. Leung EC man, Chow VC ying, Lee MK ping, Tang KP san, Li DK cheung, Lai RW man. Evaluation
of the Xpert Xpress SARS-CoV-2/Flu/RSV Assay for Simultaneous Detection of SARS-CoV-2,
Influenza A and B Viruses, and Respiratory Syncytial Virus in Nasopharyngeal Specimens.
Hayden R, editor. J Clin Microbiol. 2021 Mar 19;59(4):e02965-20.
21. Mostafa HH, Carroll KC, Hicken R, Berry GJ, Manji R, Smith E, et al. Multicenter Evaluation
of the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV Test. McAdam AJ, editor. J Clin Microbiol.
2021 Feb 18;59(3):e02955-20.
22. Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, et al. Multiplex
PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly
from clinical samples. Nat Protoc. 2017 Jun;12(6):1261–76. doi: https://doi.org/10.1038/nprot.2017.066
23. Wang L, Ng TF, Castro CJ, Marine RL, Magaña LC, Esona M, et al. Next-generation sequencing
of human respiratory syncytial virus subgroups A and B genomes. J Virol Methods. 2022 Jan;299:114335.
doi: https://doi.org/10.1016/j.jviromet.2021.114335
24. Dyrdak R, Mastafa M, Hodcroft EB, Neher RA, Albert J. Intra- and interpatient evolution
of enterovirus D68 analyzed by whole-genome deep sequencing. Virus Evol. 2019 Apr;5(1):vez007.
doi: https://doi.org/10.1093/ve/vez007
25. Hodcroft EB, Dyrdak R, Andrés C, Egli A, Reist J, García Martínez De Artola D, et
al. Evolution, geographic spreading, and demographic distribution of Enterovirus D68.
Elde NC, editor. PLOS Pathog. 2022 May 31;18(5):e1010515. doi: https://doi.org/10.1371/journal.ppat.1010515
26. Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain:
real-time tracking of pathogen evolution. Bioinformatics. 2018 Dec;34(23):4121–3.
doi: https://doi.org/10.1093/bioinformatics/bty407
27. Aksamentov I, Roemer C, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation
calling and quality control for viral genomes. J Open Source Softw. 2021;6(67):3773.
doi: https://doi.org/10.21105/joss.03773
28. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic
algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 Jan;32(1):268–74.
doi: https://doi.org/10.1093/molbev/msu300
29. Garcia-Maurino C, Brenes-Chacón H, Halabi KC, Sánchez PJ, Ramilo O, Mejias A. Trends
in age and disease severity in children hospitalized with RSV infection before and
during the COVID-19 pandemic. JAMA Pediatr. 2024 Feb;178(2):195–7. doi: https://doi.org/10.1001/jamapediatrics.2023.5431
30. Bourdeau M, Vadlamudi NK, Bastien N, Embree J, Halperin SA, Jadavji T, et al.; Canadian
Immunization Monitoring Program Active (IMPACT) Investigators. Pediatric RSV-associated
hospitalizations before and during the COVID-19 pandemic. JAMA Netw Open. 2023 Oct;6(10):e2336863–2336863.
doi: https://doi.org/10.1001/jamanetworkopen.2023.36863
31. Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, Patel SN, et al. Genetic variability
of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype
with a 72 nucleotide G gene duplication. PLoS One. 2012;7(3):e32807. doi: https://doi.org/10.1371/journal.pone.0032807
32. Trento A, Galiano M, Videla C, Carballal G, García-Barreno B, Melero JA, et al. Major
changes in the G protein of human respiratory syncytial virus isolates introduced
by a duplication of 60 nucleotides. J Gen Virol. 2003 Nov;84(Pt 11):3115–20. doi: https://doi.org/10.1099/vir.0.19357-0
33. Stein RT, Zar HJ. RSV through the COVID-19 pandemic: Burden, shifting epidemiology,
and implications for the future. Pediatr Pulmonol. 2023 Jun;58(6):1631–9. doi: https://doi.org/10.1002/ppul.26370
34. Heemskerk S, Baliatsas C, Stelma F, Nair H, Paget J, Spreeuwenberg P. Assessing the
Impact of Non-Pharmaceutical Interventions During the COVID-19 Pandemic on RSV Seasonality
in Europe. Influenza Other Respir Viruses. 2025 Jan;19(1):e70066. doi: https://doi.org/10.1111/irv.70066
35. Scholz S, Dobrindt K, Tufts J, Adams S, Ghaswalla P, Ultsch B, et al. The Burden of
Respiratory Syncytial Virus (RSV) in Germany: A Comprehensive Data Analysis Suggests
Underdetection of Hospitalisations and Deaths in Adults 60 Years and Older. Infect
Dis Ther. 2024 Aug;13(8):1759–70. doi: https://doi.org/10.1007/s40121-024-01006-0
36. Kiefer A, Pemmerl S, Kabesch M, Ambrosch A. Comparative analysis of RSV-related hospitalisations
in children and adults over a 7 year-period before, during and after the COVID-19
pandemic. J Clin Virol. 2023 Sep;166:105530. doi: https://doi.org/10.1016/j.jcv.2023.105530
37. Hönemann M, Maier M, Frille A, Thiem S, Bergs S, Williams TC, et al. Respiratory Syncytial
Virus in Adult Patients at a Tertiary Care Hospital in Germany: Clinical Features
and Molecular Epidemiology of the Fusion Protein in the Severe Respiratory Season
of 2022/2023. Viruses. 2024 Jun;16(6):943. doi: https://doi.org/10.3390/v16060943
38. Lastrucci V, Pacifici M, Puglia M, Alderotti G, Berti E, Del Riccio M, et al. Seasonality
and severity of respiratory syncytial virus during the COVID-19 pandemic: a dynamic
cohort study. Int J Infect Dis. 2024 Nov;148:107231. doi: https://doi.org/10.1016/j.ijid.2024.107231
39. Pierangeli A, Nenna R, Fracella M, Scagnolari C, Oliveto G, Sorrentino L, et al. Genetic
diversity and its impact on disease severity in respiratory syncytial virus subtype-A
and -B bronchiolitis before and after pandemic restrictions in Rome. J Infect. 2023 Oct;87(4):305–14.
doi: https://doi.org/10.1016/j.jinf.2023.07.008
40. Cutrera R, Ciofi Degli Atti ML, Dotta A, D’Amore C, Ravà L, Perno CF, et al. Epidemiology
of respiratory syncytial virus in a large pediatric hospital in Central Italy and
development of a forecasting model to predict the seasonal peak. Ital J Pediatr. 2024 Apr;50(1):65.
doi: https://doi.org/10.1186/s13052-024-01624-x
41. Redlberger-Fritz M, Springer DN, Aberle SW, Camp JV, Aberle JH. Respiratory syncytial
virus surge in 2022 caused by lineages already present before the COVID-19 pandemic.
J Med Virol. 2023 Jun;95(6):e28830. doi: https://doi.org/10.1002/jmv.28830
42. Rios-Guzman E, Simons LM, Dean TJ, Agnes F, Pawlowski A, Alisoltanidehkordi A, et
al. Deviations in RSV epidemiological patterns and population structures in the United
States following the COVID-19 pandemic. Nat Commun. 2024 Apr;15(1):3374. doi: https://doi.org/10.1038/s41467-024-47757-9
43. de Korne-Elenbaas J, Rimaite A, Topolsky I, Dreifuss D, Bürki C, Fuhrmann L, et al. Wastewater-based
sequencing of Respiratory Syncytial Virus enables tracking of lineages and identifying
mutations at antigenic sites. medRxiv [Preprint]. 2025 medRxiv [cited 2025 Mar 7].
p. 2025.02.28.25321637. Available from: https://www.medrxiv.org/content/10.1101/2025.02.28.25321637v1 10.1101/2025.02.28.25321637
44. Yunker M, Fall A, Norton JM, Abdullah O, Villafuerte DA, Pekosz A, et al. Genomic
Evolution and Surveillance of Respiratory Syncytial Virus during the 2023-2024 Season.
Viruses. 2024 Jul;16(7):1122. doi: https://doi.org/10.3390/v16071122
45. de Jesus-Cornejo J, Hechanova-Cruz RA, Sornillo JB, Okamoto M, Oshitani H; Viral Etiology
Working Group. Prolonged RSV circulation in 2022 to 2023 associated with the emergence
of a novel RSV-B clade in Biliran, Philippines. J Infect. 2025 Apr;90(4):106467. doi: https://doi.org/10.1016/j.jinf.2025.106467
46. Jafri HS, Wu X, Makari D, Henrickson KJ. Distribution of respiratory syncytial virus
subtypes A and B among infants presenting to the emergency department with lower respiratory
tract infection or apnea. Pediatr Infect Dis J. 2013 Apr;32(4):335–40. doi: https://doi.org/10.1097/INF.0b013e318282603a
47. Midulla F, Nenna R, Scagnolari C, Petrarca L, Frassanito A, Viscido A, et al. How
respiratory syncytial virus genotypes influence the clinical course in infants hospitalized
for bronchiolitis. J Infect Dis. 2019 Jan;219(4):526–34. doi: https://doi.org/10.1093/infdis/jiy496
48. Jung JA, Wi PH, Kim H. Kim H sol. Comparisons of clinical characteristics between
Respiratory Syncytial Virus A and B infection. J Allergy Clin Immunol. 2020;145(2):AB131.
doi: https://doi.org/10.1016/j.jaci.2019.12.520
49. Vos LM, Oosterheert JJ, Kuil SD, Viveen M, Bont LJ, Hoepelman AI, et al. High epidemic
burden of RSV disease coinciding with genetic alterations causing amino acid substitutions
in the RSV G-protein during the 2016/2017 season in The Netherlands. J Clin Virol.
2019 Mar;112:20–6. doi: https://doi.org/10.1016/j.jcv.2019.01.007
50. Irwin KK, Renzette N, Kowalik TF, Jensen JD. Antiviral drug resistance as an adaptive
process. Virus Evol. 2016 Jun;2(1):vew014. doi: https://doi.org/10.1093/ve/vew014
51. Gosert R, Leuzinger K, Kuznetsov A, Neher RA. Respiratory syncytial virus A amplicon
sequencing dataset, Basel, Switzerland, 2021-2024 [dataset]. 2025 Jun 16 [cited 2025
Sep 23]. Pathoplexus. Available from: 10.62599/PP_SS_201.2
52. Gosert R, Leuzinger K, Kuznetsov A, Neher RA. Respiratory syncytial virus B amplicon
sequencing dataset, Basel, Switzerland, 2020-2024 [dataset]. 2025 Jun 16 [cited 2025
Sep 23]. Pathoplexus. Available from: 10.62599/PP_SS_202.2
Appendix
The appendix is available in the PDF version of the article and the supplementary
files are available for download as separate files at https://doi.org/10.57187/s.4600.



