Rapid detection of the source of a Listeria monocytogenes outbreak in Switzerland
through routine interviewing of patients and whole-genome sequencing
DOI: https://doi.org/https://doi.org/10.57187/s.3745
Cornelia
Speichab,
Roger
Stephanc,
Nicola
Dhimad,
Florian Hollensteine,
Jule
Horlbogc,
Giulia
Delventoab,
Ekkehardt
Altpeterd,
Meike
Zuskeab,
Michelle
Raessd,
Helena
Greterab
a
Swiss Tropical and Public Health Institute, Allschwil, Switzerland
b University of Basel, Basel, Switzerland
c
Institute for Food Safety and Hygiene, National Reference
Laboratory for Enteropathogenic Bacteria and Listeria (NENT), Vetsuisse Faculty,
University of Zurich, Zurich, Switzerland
d Federal Office of Public Health, Liebefeld, Bern, Switzerland
e Kantonales Laboratorium Thurgau, Frauenfeld,
Switzerland
Summary
AIMS OF THE
STUDY: Listeriosis is a notifiable disease in Switzerland.
In summer 2022, the Swiss Federal Office of Public Health noticed an increase in
reports of listeriosis cases, indicating a possible ongoing outbreak. Here we present
the approaches applied for rapidly confirming the outbreak, detecting the underlying
source of infection and the measures put in place to eliminate it and contain the
outbreak.
METHODS: For close surveillance and early detection of outbreak situations with
their possible sources, listeriosis patients in Switzerland are systematically interviewed
about risk behaviours and foods consumed prior to the infection. Listeria monocytogenes isolates derived from
patients in medical laboratories are sent to the National Reference Laboratory for
Enteropathogenic Bacteria and Listeria, where they routinely undergo whole-genome
sequencing. Interview and whole-genome sequencing data are continuously linked for
comparison and analysis.
RESULTS: In summer 2022, 20 patient-derived L. monocytogenes serotype
4b sequence type 388 strains were found to belong to an outbreak cluster (≤10 different
alleles between neighbouring isolates) based on core genome multilocus sequence
typing analysis. Geographically, 18 of 20 outbreak cases occurred in northeastern
Switzerland. The median age of patients was
77.4 years (range: 58.1–89.7), with both sexes equally affected. Rolling analysis
of the interview data revealed smoked trout from a local producer as a suspected
infection source, triggering an on-site investigation of the production facility
and sampling of the suspected products by the responsible
cantonal food inspection team on 15 July 2022. Seven of ten samples tested positive
for L. monocytogenes and the respective cantonal authority ordered
a ban on production and distribution as well as a product recall. The Federal Food
Safety and Veterinary Office released a nationwide public alert covering the smoked
fish products concerned. Whole-genome sequencing analysis confirmed the interrelatedness
of the L. monocytogenes smoked trout product
isolates and the patient-derived isolates. Following the ban on production and distribution
and the product recall, reporting of new outbreak-related cases rapidly dropped
to zero.
CONCLUSIONS: This listeriosis outbreak could be contained within a relatively short
time thanks to identification of the source of contamination through the established
combined approach of timely interviewing of every listeriosis patient or a representative
and continuous molecular analysis of the patient- and food-derived L. monocytogenes isolates. These findings
highlight the effectiveness of this well-established, joint approach involving the
federal and cantonal authorities and the research institutions mandated to contain
listeriosis outbreaks in Switzerland.
Introduction
Listeriosis is a rare disease in Switzerland
with 40 to 60 confirmed cases every year in a population of about 8.7 million inhabitants
[1]. However, the disease may be severe and
life-threatening, especially for immunocompromised and elderly people [1–4]. Due to
its high case fatality rate, listeriosis
ranks among the leading causes of death due to food-borne illness and
consequently has a significant negative impact on public health and the economy
in most European countries [3, 4]. Symptoms
of listeriosis may be mild such as diarrhoea and low-grade fever, but the disease
can also lead to severe symptoms including sepsis and organ damage [2, 5]. Furthermore,
Listeria monocytogenes has the ability to cross the blood-brain barrier
leading to meningitis and brain infection [5]. In pregnant women, L. monocytogenes may cross the placental
barrier leading to diaplacental infections of the foetus with high risk of subsequent
abortion [6]. The long incubation period of approximately 11 days (10% of cases
showing incubation periods >4 weeks) [7]
is a major challenge when searching for the source of infection with listeriosis
patients.
Given their ubiquitous occurrence in the environment
including the soil and/or surface water, L.
monocytogenes can be present in raw foods (e.g. meat, fish, poultry, vegetables,
raw milk) [8].
Since 1974, human listeriosis is classified
as a notifiable disease in Switzerland and hence, every culture- or polymerase chain
reaction (PCR)-confirmed human listeriosis case has to be reported to the Swiss Federal
Office of Public Health (SFOPH) [9]. Additionally, all isolates are sent to the
National Reference Laboratory for Enteropathogenic Bacteria and Listeria (NENT), where
all clinical L. monocytogenes isolates
have been routinely whole-genome sequenced since August 2018. This procedure ensures
early detection of L. monocytogenes clusters,
which indicate that several patients are suffering from an infection with the
same strain. Furthermore, for close surveillance and rapid detection of potential
sources of contamination to limit outbreaks, SFOPH has commissioned the Competence
Centre for Epidemiological Outbreak Investigations
(KEA) at the Swiss Tropical and Public Health Institute (Swiss TPH) to routinely interview
all patients diagnosed and reported with listeriosis using a standardised questionnaire.
Topics covered in these interviews include, among others, exposure risk behaviours
and food consumption prior to illness. Here, we describe the processes leading
to the detection of a regional listeriosis outbreak in summer 2022 linked to L. monocytogenes serovar 4b, sequence type
388, and the intervention measures taken to contain the outbreak.
Materials and methods
Data collection
In accordance with the standard procedure for
listeriosis surveillance in Switzerland, isolates of (patient) samples that tested
positive for L. monocytogenes at different
medical laboratories were routinely sent to NENT for molecular analysis (routine
whole-genome sequencing). Similarly, the attending physician of each patient reported
to SFOPH was routinely contacted by KEA and asked for his/her assessment of the
patient’s ability to be interviewed. If a direct interview was ruled out due to
the patient’s health status, the physician was asked to identify a proxy person
(relative, partner, caregiver, etc). In rare cases, the physician advised against
conducting an interview. Reasons for advising against an interview included language
barriers (neither the patient nor the proxy person could speak or understand German,
French, Italian or English), non-availability or absence of a proxy person, or consideration
for the potential proxy person in light of the patient’s serious illness or recent
death. Attending physicians were also interrogated regarding their information and/or
suspicions concerning the source of infection, if any. Next, an information letter
was sent to every eligible patient or his/her representative, announcing the anticipated
telephone interview. A trained interviewer then called the person. In this call,
the interviewer sought oral informed consent after informing patients that participation
is voluntary, confidentiality is guaranteed, and that they do not have to
provide answers if they are not willing to do so and this would not have any negative
consequences for either themselves or their representative. Also, patients were
informed that by sharing detailed information such as the names of a brand, store,
producer or restaurant, they were not informing against anyone but might help to
identify a problem fast and to prevent harm to both the business and to public health.
The telephone interview took approximately 20–30 minutes. The standardised questionnaire
contained the following categories and covered a time frame of four weeks prior
to symptom onset, a period in which incubation of 90% of the cases can be assumed
[7]: (a) national and international travel; (b) out-of-home food consumption in
restaurants, canteens or take-aways; (c) special diets, food allergies and intolerances;
(d) habitual shopping places for different food categories; and (e) habitual and
recent consumption of different food categories (fish and seafood, meat and sausages,
poultry, cheese and dairy products, vegetables, fruits, salads and ready-to-eat
food items). For grocery shopping and food consumption, habitual practices were
taken into account because of the difficulty in remembering food items consumed
over a time period that is necessarily long in the case of listeriosis (incubation
period is median 11 days and >4 weeks in up to 10% of cases).
Data management
During the interview, data were recorded on
paper and then transcribed to a digital database (Microsoft Access 2016). Anonymisation
was achieved through the strict separation of personal identifying data and the
interview data, whereas all paper-based or digital documentation of personal identifiers
were stored separately and secured. Both access to files and to the database were
restricted to the team members of SFOPH and KEA involved in this study by physical
locks and digital login protection, respectively, as per Swiss federal data protection
laws. The whole-genome sequencing results for each case were shared by NENT in an
anonymised form (Microsoft Excel 2016) every week, and matching of surveillance
and whole-genome sequencing results was performed on sample identification number
and date of birth. Data were inspected case-by-case and compared with other cases
of the respective cluster on a rolling basis in order to allow identification
of similarities in travel and food consumption habits as well as consumed food items
between cluster-associated cases.
Statistical analysis
Descriptive statistical analysis was performed
using Microsoft Excel 2016 and R version 4.1.3 and summary statistics are presented
as median (interquartile range), counts and percentages.
Laboratory work-up
For the molecular analysis of all L. monocytogenes
isolates, routine whole-genome sequencing on human, food and factory environmental
strains was performed using Illumina MiSeq next-generation sequencing technology
(Illumina, San Diego, CA, USA) as described in [10].
Sequencing reads were mapped against a multilocus sequence typing (MLST) scheme
based on seven housekeeping genes and a 1701-locus core genome multilocus sequence
typing (cgMLST) scheme using Ridom SeqSphere+ software version 7.7.5. Strain types
and cluster types were determined upon submission
to the L. monocytogenes cgMLST Ridom SeqSphere+ server (http://www.cgmlst.org/ncs/schema/690488/).
Clusters are defined as a group of isolates with ≤10 different alleles between neighbouring
isolates [10].
Based on the results of the patient interviews,
food products of a local producer were collected by the responsible cantonal authorities
(Kantonales Laboratorium Thurgau) in July 2022 and processed according to ISO 11290-1
and ISO 11290-2. Moreover, 60 swabs from the production environment of the respective
facility were collected. Swabs were incubated in Half Frazer Broth (HFB, BioRad,
Cressier, Switzerland) at 30 °C for 48 hours. L. monocytogenes was detected
by real-time PCR using the Assurance Genetic Detection System (GDS®, Endotell, Allschwil,
Switzerland) according to the manufacturer’s instructions. To obtain strains for
whole-genome sequencing, the PCR-positive HFB cultures were streaked on Oxoid chromogenic
Listeria agar (OCLA) plates (Oxoid, Pratteln, Switzerland) and incubated
at 37 °C for 48 hours.
Ethical considerations
The procedures involved in the routine interviewing
of all listeriosis patients in Switzerland through standardised questionnaires for
surveillance are authorised by the Swiss Epidemics Act (SR818.101, EpG) and hence
no additional ethical clearance was needed for this subanalysis. All included patients
were asked for their oral informed consent before the interview was conducted. All
data were stored, managed and analysed in anonymised form.
Results
Outbreak progression
In the third week of April 2022, four cases
of listeriosis were reported to SFOPH, all living in the canton of St. Gallen in
northeastern Switzerland. This represented an unusual accumulation of listeriosis
cases in the canton as it had reported only one listeriosis case earlier in 2022
(in January). Whole-genome sequencing analyses confirmed the interrelatedness between
two of the four reported cases (L. monocytogenes serotype 4b sequence type
388 cgMLST 18052) and data from interviewing these two patients in early May 2022
revealed that both patients reported having consumed fish (without yet clear congruency
among the mentioned fish species or products) bought at local stores or farmers’
markets.
Another listeriosis case in St. Gallen diagnosed
seven weeks later was linked to the same cluster based on whole-genome
sequencing analysis. In the same period, four more listeriosis cases from other
regions in Switzerland had been reported, with no relation to the cluster. However,
the third cluster-associated case in St. Gallen marked the starting point of a strong
increase in cases reported from northeastern Switzerland for the next month, with
11 additional cases linked to the cluster, resulting in a total of 14 cases to mid-July
2022 when corrective measures could be taken (figure 1).
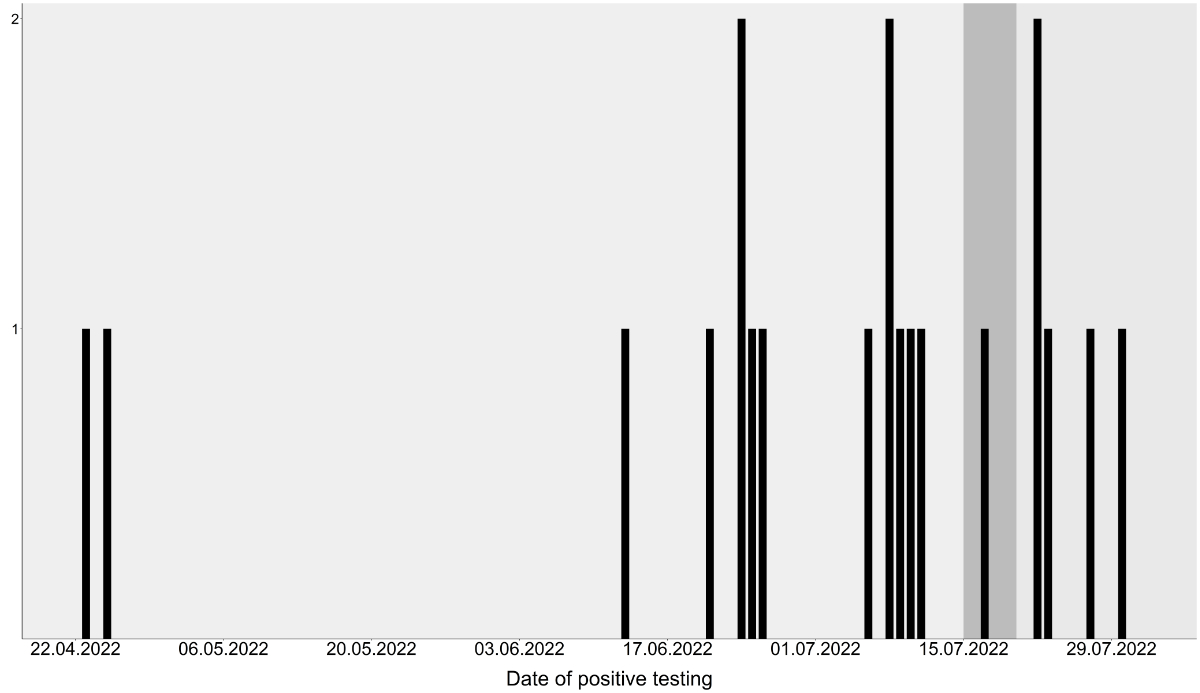
Figure 1Occurrence over time of listeriosis cases
related to the smoked trout outbreak in Switzerland. Corrective measures
started on 15 July 2022 and the source of contamination identified and
respective products recalled on 20 July 2022. Respective dates indicated by
changing background colours.
After implementation of corrective measures
on 15 July 2022, an additional six listeriosis patients were identified as belonging
to the outbreak cluster, with the last related patient testing positive on 30 July
2022 (figure 1).
Outbreak investigation
Interviewing of the patients took place in a
rolling procedure following the standardised questionnaire, but with special probing
on fish consumption during the four weeks prior to symptoms onset as per the suspicion
from the first two interviews. Out of the 14 cluster-associated listeriosis cases
diagnosed to mid-July 2022, 8 could be interviewed between 6 May 2022 and 15 July
2022 (table 1). All patients or their respective proxy people interviewed before
15 July 2022 confirmed fish consumption during the critical period (figure 2). Six
of the interviewed or proxy-interviewed patients reported having consumed smoked
trout during the critical infection period. Among them, four patients named the
same producer for the consumed smoked trout, with the first mention of this specific
product recorded on 4 July 2022 in two independent interviews, and confirmation
of the suspicion by two other patients / proxies by 15 July 2022.
Table 1Overview of the outbreak interviewing process. Of 20 patients, six were not interviewed
because (a) the patient did not answer the phone despite several attempts (n = 1),
(b) the attending physician advised against contacting the patient’s proxy
person out of consideration for their partner’s severe illness following the
listeriosis infection (n = 1) and (c) patients diagnosed only after
identification of the source of contamination and/or patients already
mentioning consumption of smoked trout to their attending physician (n = 4).
| Direct
interviews, n (%) |
8 (40%) |
| Interviews
before 15 July 2023 and thus contributing to identification of the source of
contamination, n (%) |
5 (25%) |
| Proxy interviews, n (%); proxy people were the patient’s spouse
(n = 5) or daughter (n = 1) |
6 (30%) |
| Proxy
interviews before 15 July 2023 and thus contributing to identification of the
source of contamination, n (%) |
3 (15%) |
| Time from
diagnosis to interview in days, median (interquartile range) |
10 (9–15) |
| Time from
diagnosis to physician interview in days, median (interquartile range) |
7 (5–11) |

Figure 2Source of infection identification through
eight cluster patient interviews leading to corrective measures from 15 July
2022. * Includes one patient mentioning a slightly
different name of the smoked trout producer that was assumed to be a wrong
recombination of letters in the patient’s memory in the course of the
investigation.
After on-site audit and sampling on 15 July 2022, additional 2 of the listeriosis
cases reported before that date, as well as 4 cases reported thereafter could still
be interviewed (among which 3 proxy interviews) as per routine practice, to
further allow acting as soon as possible before final whole-genome sequencing confirmation
was available.
Outbreak management
Based on the repeated reporting of consumption
of smoked trout from the same producer, the responsible cantonal authorities were
informed of the suspicion on Friday 15 July 2022. On the same day, a first audit
of the production facility and sampling of two of the suspected products was initiated.
Smoked trout products sampled during the on-site investigation were analysed in
the cantonal laboratory and seven of ten samples tested positive for L. monocytogenes. In six samples, L. monocytogenes was qualitatively detectable
in 25 g, while in one sample 10 colony-forming units/gram were detected.
The producer company supplied several of the
main retailers in the respective area with their products, ran a farm shop and sold
the products directly to the public at a weekly market and online. After detection
of L. monocytogenes in the sampled smoked
trout, the respective cantonal authority ordered a ban on production and distribution
as well as a product recall on 20 July 2022. To alert the public, the Federal Food
Safety and Veterinary Office released a nationwide public warning covering the implicated
products on 21 July 2022. Finally, following the whole-genome sequencing confirmation
of the interrelatedness of the L. monocytogenes
isolates from the smoked trout samples and the patient-derived isolates on 23 July
2023 (figure 3), the authorities issued comprehensive measures on 26 July 2022.
These included extensive sanitation measures at the manufacturing facility informed
by the analysis of 60 swabs from the production environment and a revision of the
company’s self-monitoring policy. After the company took action to restore its
legal status and listeria was no longer detectable in environmental samples and
ready-to-eat products, the ban on production and distribution was lifted several
months later.
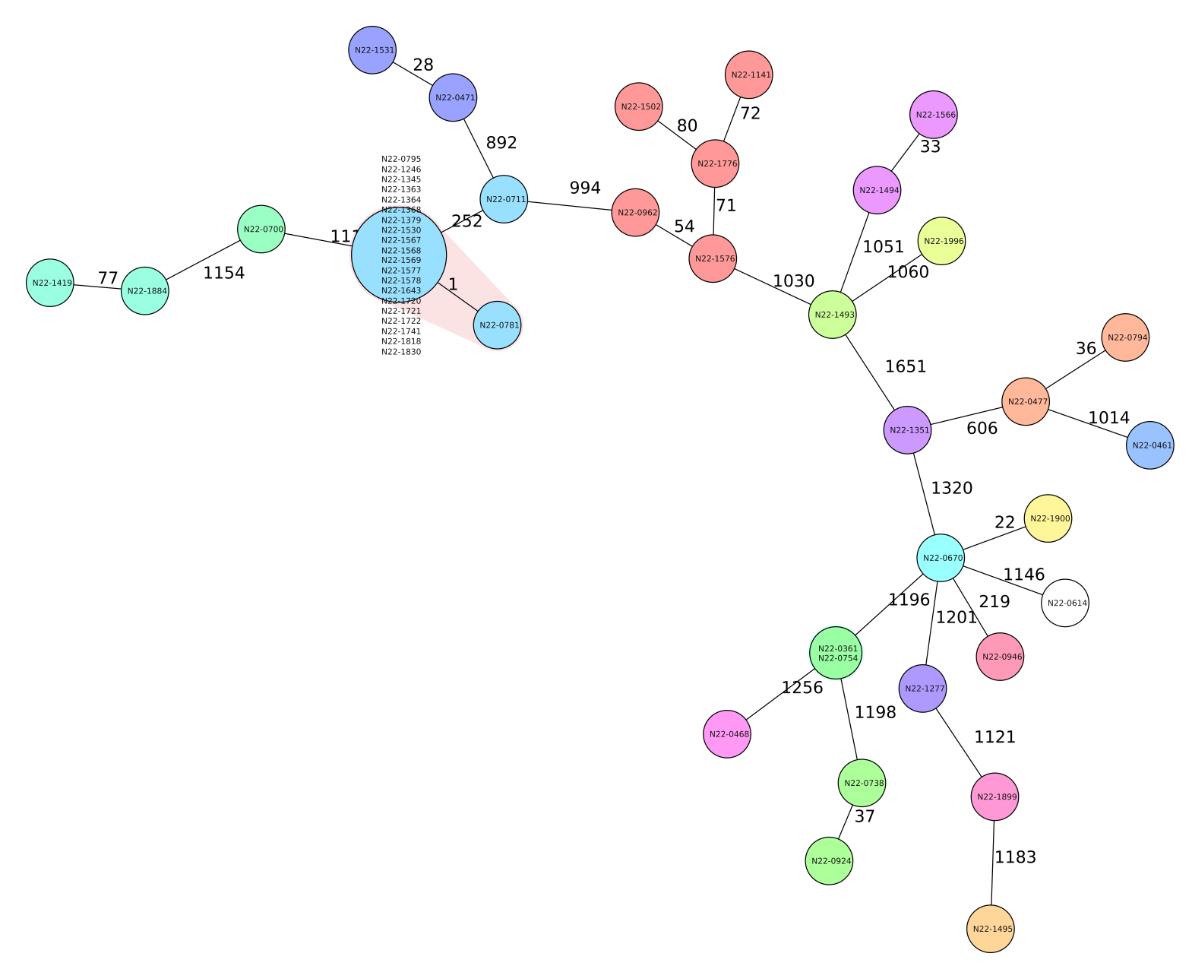
Figure 3Minimum-spanning tree based on cgMLST allelic
profiles of isolates from all diagnosed human Listeria monocytogenes cases in Switzerland between 1 March 2022
and 31 August 2022 as well as a representative smoked trout sample collected at
the production facility on 15 July 2022. Each circle represents an allelic
profile based on sequence analysis of 1701 cgMLST target genes. Values on
connecting lines indicate number of allelic differences between two strains.
The food isolate only differs from the outbreak strain in one allele as
highlighted by red shading.
Characteristics of the outbreak patients
The 20 outbreak cases were reported from six
Swiss cantons. Demographic characteristics of affected patients are shown in table
2. No clinical characteristics are collected in a standardised manner in routine
listeriosis surveillance given that rapid identification and elimination of the
source of infection are the main objectives in the case of food-borne disease outbreak
investigations. Clinical characteristics reported to SFOPH via clinical notifications
from attending physicians are not routinely shared with either KEA or NENT and death
certifications are not routinely shared
with SFOPH by hospitals, except if directly related to the disease case and not
to comorbidities as per the attending physician’s judgement.
Table 2Characteristics of the 20 outbreak
patients.
| Age in years; median
(interquartile range): range |
77.4 (67.2–82.5) 58.1–89.7 |
| Male, n (%) |
10 (50%) |
The outbreak patient samples that tested positive
for L. monocytogenes at different medical
laboratories were blood (17), pleura (1), cerebrospinal fluid (1) and stool (1)
samples. The analysed patient isolates did not differ by any allele while the analysed
food isolates differed from the patient isolates by one allele (figure 3).
In total, 14 of 20 outbreak-associated patients
were interviewed in the context of this listeriosis outbreak investigation, while
whole-genome sequencing typing was available for all 20 patients. As per standard
procedures, attending physicians could be contacted for 19 of the 20 affected patients
(for one patient the attending physician could not be reached, so the interview
was conducted directly with the patient without pre-discussion with the attending
physician). For 3 of the 6 patients who could not be interviewed personally or through
a proxy person by the KEA team, the attending physician confirmed that during
questioning the patient remembered having consumed smoked trout from the affected
producer (figure 4). Thus in total, 17 of the 20 L. monocytogenes outbreak-associated patients mentioned – personally, through their
representative or through their attending physician – consumption of fish. Consumption
of smoked trout was confirmed by/for 15 patients and 9 patients mentioned the same
producer for the smoked trout that they had consumed (figure 4). One of the early
detected cases in this outbreak mentioned a smoked trout producer with a similar
but not identical name. Furthermore, two of the later listeriosis cases mentioned
having bought their smoked trout at specialist gourmet food distributors that
list smoked trout from the affected producer in their product catalogue. Table 1
characterises the conducted interviews in terms of who was interviewed and within
what timeframe.

Figure 4Source of infection identification by the 20
patients affected by listeriosis from the smoked trout outbreak in Switzerland
in summer 2022. * Includes two patients only mentioning smoked trout from a gourmet
food reseller that clearly lists smoked trout from the affected producer as a
supplier and one patient mentioning a slightly different name of the smoked
trout producer that was assumed to be a wrong recombination of letters in the
patient’s memory in the course of the investigation.
Discussion
Rapid identification and elimination of the
source of infection is the main objective in investigations of food-borne disease
outbreaks. Epidemiological surveillance combined with molecular analysis of the
pathogens have proven highly effective for early detection and containment of food-borne
disease outbreaks. Epidemiological surveillance through timely routine interviewing
of all listeriosis patients in Switzerland played a key role in rapidly identifying
the source of contamination and provided the basis for re-establishing food safety
for consumers in this outbreak. Through the routine characterisation of all positive
human-derived L. monocytogenes isolates
by the Swiss reference laboratory NENT and routine investigation of food habits
of all listeriosis patients through interviews based on a standardised questionnaire,
similarities in food habits and food choices could be analysed from early on. Fish
was suspected as a potential source of contamination very early in this outbreak
based on the reports by the first two patients of the cluster mentioning repeated
fish consumption and purchase of fish at markets and farm shops. This observation
was taken up and responded to by integrating additional, targeted probing on fish
consumption into interviews with all later patients. However, the type of fish or
fish product remained obscure for the following weeks with the first three patients
mentioning consumption of different fish species and products, and smoked trout
consumption only mentioned by one of the first three patients. Subsequent interviews
with patients four to eight revealed overlapping information on consumption of smoked
trout, the brand, producer and retailer where smoked trout had been purchased, and
hence provided the decisive information that led to the targeted audit of the suspected
production facility.
Smoked fish products such as smoked salmon and
smoked trout are ranked as high-risk products for L. monocytogenes infection given their high pH and aw value,
combined with the fact that they are generally consumed directly as the product
does not require any cooking processes and is more of a ready-to-eat item [11, 12].
A recent study from Germany investigated
the interrelatedness between clinical L. monocytogenes
isolates and non-clinical isolates from fish and other food samples and found high
correspondences [13]. Reasons for contamination
might be either primary contamination of the fish itself or cross-contamination
within the manufacturing and retailing process up to consumption [2]. In 2020, of
12 larger L. monocytogenes outbreaks reported in 34 European countries, eight
outbreaks have been attributed to consumption of ready-to-eat fish products, two
to consumption of meat products and two to consumption of milk and dairy products
[14–16], emphasising the high risk of listeriosis
related to the consumption of ready-to-eat fish products. Also, in a large analysis
collecting more than 800 samples from the production environment and fresh fish
from two processing facilities producing smoked fish, Hoffman et al. identified drains
as the locations
carrying the highest load of L. monocytogenes,
if any, although product contamination can occur at various steps throughout the
production line and any following handling of the product [17].
In the present outbreak, related cases were
reported over a period of nearly four months despite the short shelf life of the
food item identified. This is an indication of a persistent contamination source
in the production facility rather than contamination of a specific batch. Trout
bred at the same facility and intended for sale as fresh fish was not connected
to any listeriosis cases and had tested negative in the analysis carried out.
Clear limitations of this study are the pragmatically
decided, long yet potentially still insufficient recall period of four weeks prior
to symptom onset used in routine listeriosis surveillance in Switzerland as well
as the lack of systematic analysis of clinical symptoms. Similarly, the practice
of obtaining information from family representatives or even attending doctors when
it could not be obtained directly from patients might mean that important information
is missed but it remains the best alternative solution for getting information.
However, the combined approach (patient interviews and whole-genome
sequencing) and the close collaboration of cantonal and national authorities with
mandated research institutions have proven effective and rapid in numerous listeriosis
outbreaks in the past.
The parties involved in listeriosis outbreak
investigations in Switzerland are in constant communication, allowing for continual
revision and improvement of their approaches. In order to further reduce the risk
of listeriosis infections for consumers, one important pillar of the current Federal
Act on Foodstuffs and Utility Articles (Foodstuff Act, FSA; SR: 817.0) [18] is the
existing self-monitoring by, official audits of and awareness-raising among food-producing
companies. A second pillar is raising
awareness among vulnerable population groups, such as the elderly, immunosuppressed
people and pregnant women, about food items with an increased risk for L. monocytogenes contamination.
Data sources and availability
The sequence data of a representative strain
of the outbreak (N22-1530) has been deposited at the NCBI BioSample database under
project number SAMN32990935, accession number JAQQAV000000000.
Acknowledgments
We thank all participants for their willingness
to participate in an interview during a time of physical illness. Further, we thank
all the attending physicians for their information sharing and the authorities of
the cantons Thurgau, St. Gallen, Vaud and Zurich and the Federal Office of Public
Health and Federal Food Safety and Veterinary Office, for the good and uncomplicated
collaboration and M. Stevens for the bioinformatic support.
Helena Greter
Swiss TPH
Kreuzstrasse
2
CH-4123 Allschwil
helena.greter[at]swisstph.ch
References
1 Bundesamt für Gesundheit (BAG). Listeriose - Jährliche Fallmeldungen und Inzidenzen
der letzten 10 Jahre und aktuelles Jahr bis Woche 39/2022 https://www.bag.admin.ch/bag/de/home/zahlen-und-statistiken/zahlen-zu-infektionskrankheiten.exturl.html/aHR0cHM6Ly9tZWxkZXN5c3RlbWUuYmFnYXBwcy5jaC9pbmZyZX/BvcnRpbmcvZGF0ZW5kZXRhaWxzL2QvbGlzdGVyaWEuaHRtbD93/ZWJncmFiPWlnbm9yZQ==.html [updated 04.10.2022].
2 Krämer J, Prange A. Lebensmittel-Mikrobiologie. 7th ed. Stuttgart: Utb GmbH; 2016.
3 Werber D, Hille K, Frank C, Dehnert M, Altmann D, Müller-Nordhorn J, et al. Years
of potential life lost for six major enteric pathogens, Germany, 2004-2008. Epidemiol
Infect. 2013 May;141(5):961–8. 10.1017/S0950268812001550
4 Barton Behravesh C, Jones TF, Vugia DJ, Long C, Marcus R, Smith K, et al.; FoodNet
Working Group. Deaths associated with bacterial pathogens transmitted commonly through
food: foodborne diseases active surveillance network (FoodNet), 1996-2005. J Infect
Dis. 2011 Jul;204(2):263–7. 10.1093/infdis/jir263
5 Malinverni R, Bille J, Perret C, Regli F, Tanner F, Glauser MP. Listériose épidémique.
Observation de 25 cas en 15 mois au Centre hospitalier universitaire vaudois. Schweiz
Med Wochenschr. 1985 Jan;115(1):2–10.
6 Lamond NM, Freitag NE. Vertical Transmission of Listeria monocytogenes: Probing the Balance between Protection from Pathogens and Fetal Tolerance. Pathogens.
2018 May;7(2):52. 10.3390/pathogens7020052
7 Angelo KM, Jackson KA, Wong KK, Hoekstra RM, Jackson BR. Assessment of the Incubation
Period for Invasive Listeriosis. Clin Infect Dis. 2016 Dec;63(11):1487–9. 10.1093/cid/ciw569
8 Vivant AL, Garmyn D, Piveteau P. Listeria monocytogenes, a down-to-earth pathogen. Front Cell Infect Mi; 2013. p. 3.
9 Baumgartner A, Schmid H. Listeria monocytogenes in genussfertigen Lebensmitteln: eine Auswertung der amtlichen Untersuchungen in
der Schweiz der Jahre 2006-2008. Journal für Verbraucherschutz und Lebensmittelsicherheit
/ Journal of Consumer Protection and Food Safety. 2013;8:109-17.
10 Nüesch-Inderbinen M, Bloemberg GV, Müller A, Stevens MJ, Cernela N, Kollöffel B, et
al. Listeriosis Caused by Persistence of Listeria monocytogenes Serotype 4b Sequence Type 6 in Cheese Production Environment. Emerg Infect Dis. 2021 Jan;27(1):284–8.
10.3201/eid2701.203266
11 U.S. Food & Drug Administration. Water Activity (aw) in Foods https://www.fda.gov/inspections-compliance-enforcement-and-criminal-investigations/inspection-technical-guides/water-activity-aw-foods2014
12 Kiczorowska B, Samolińska W, Grela ER, Bik-Małodzińska M. Nutrient and Mineral Profile
of Chosen Fresh and Smoked Fish. Nutrients. 2019 Jun;11(7):1448. 10.3390/nu11071448
13 Lachmann RA-O, Halbedel SA-O, Lüth SA-O, Holzer A, Adler M, Pietzka A, et al. Invasive
listeriosis outbreaks and salmon products: a genomic, epidemiological study. (2222-1751
(Electronic)).
14 European Food Safety Authority (EFSA). Foodborne outbreaks - dashboard https://www.efsa.europa.eu/de/microstrategy/FBO-dashboard2020
15 European Food Safety Authority; European Centre for Disease Prevention and Control.
The European Union One Health 2020 Zoonoses Report. EFSA J. 2021 Dec;19(12):e06971.
16 Halbedel S, Sperle I, Lachmann R, Kleta S, Fischer MA, Wamp S, et al. Large multi-country
outbreak of invasive listeriosis by a Listeria monocytogenes ST394 clone linked to smoked rainbow trout, 2020-2021 [in revision]. Microbiol Spectr.
2023 Jun;11(3):e0352022. 10.1128/spectrum.03520-22
17 Hoffman AD, Gall KL, Norton DM, Wiedmann M. Listeria monocytogenes contamination patterns for the smoked fish processing environment and for raw fish.
J Food Prot. 2003 Jan;66(1):52–60. 10.4315/0362-028X-66.1.52
18 Swiss Federal Act on Foodstuffs and Utility Articles, Stat. SR 817.0 (20Jun2014, 2014).



