Association between the number of symptomatic mpox cases and the detection of mpox
virus DNA in wastewater in Switzerland: an observational surveillance study
DOI: https://doi.org/https://doi.org/10.57187/s.3706
Claudia Baguttia,
Monica Alt Huga,
Philippe Heima,
Evelyn Ilg Hampea,
Philipp Hübnera,
Timothy R. Julianb,
Katrin N. Kochc,
Kerstin Grosheintzd,
Melanie Krausd,
Carla Schaubhutd,
Rahel Tarnutzerd,
Eva Würfeld,
Simon Fuchsd,
Sarah Tschudin-Suttere
a State
Laboratory of Basel-Stadt, Basel, Switzerland
b Eawag,
Swiss Federal Institute of Aquatic Science and Technology, Dübendorf,
Switzerland
c Cantonal
Office of Public Health, Department of Economics and Health, Canton of
Basel-Landschaft, Liestal, Switzerland
d Department
of Health, Canton of Basel-Stadt, Basel, Switzerland
e Division
of Infectious Diseases & Hospital Epidemiology, University Hospital Basel,
Switzerland / University of Basel, Basel, Switzerland
Summary
AIM OF THE STUDY: The COVID-19 pandemic has drawn attention to the benefit of
wastewater-based epidemiology, particularly when case numbers are
underreported. Underreporting may be an issue with mpox, where biological
reasons and stigma may prevent patients from getting tested. Therefore, we aimed
to assess the validity of wastewater surveillance for monitoring mpox virus
DNA in wastewater of a Central European city and its association with official
case numbers.
METHODS:
Wastewater samples were collected between 1 July and 28 August 2022 in the
catchment area of Basel, Switzerland, and the number of mpox virus genome
copies they contained was determined by real-time quantitative PCR. Logistic
regression analyses were used to determine the odds of detectability of mpox
virus DNA in wastewater, categorised as detectable or undetectable. Mann–Whitney
U tests were used to determine associations between samples that tested
positive for the mpox virus and officially reported cases and patients’ recorded
symptomatic phases.
RESULTS: Mpox virus DNA was detected in 15 of 39 wastewater samples. The number of
positive wastewater samples was associated with the number of symptomatic cases
(odds ratio [OR] = 2.18, 95% confidence interval (CI) = 1.38–3.43, p = 0.001). The
number of symptomatic cases differed significantly between days with positive versus
negative wastewater results (median = 11 and 8, respectively, p = 0.0024).
CONCLUSION: Mpox virus DNA was detectable in wastewater, even when officially reported
case numbers were low (0–3 newly reported mpox cases corresponding to 6–12
symptomatic patients). Detectability in wastewater was significantly associated
with the number of symptomatic patients within the catchment area. These
findings illustrate the value of wastewater-based surveillance systems when
assessing the prevalence of emerging and circulating infectious diseases.
Introduction
Mpox has been described as an endemic zoonotic disease in Western and Central Africa
since the 1970s [1]. Individual cases and
small outbreaks outside the endemic regions were import- and travel-related and
did not persist. In May 2022, a novel mpox clade (IIb [2, 3]) emerged in non-endemic
countries,
mainly in Europe. Since mpox clade IIb is primarily transmitted sexually, it
is likely associated with stigma. Therefore, hesitancy to get tested may impair
official reporting of mpox infections [4].
Furthermore, case numbers are likely underestimated due to the disease’s non-specific
symptoms (especially at disease
onset), such as fever, myalgia, fatigue, and headache, asymptomatic course [5], and
long
incubation time of up to 21 days [6]. Therefore,
independent and unbiased surveillance systems are needed to estimate the true prevalence
of mpox cases.
Studies
from the US, France, Italy, Spain, and the Netherlands [7–11] reported successfully
detecting mpox virus DNA in
wastewater and identifying the clade [12].
Amongst these recently published studies, two compared their wastewater data with
mpox case data but did not perform correlation analyses [7, 10]. In their most recent
study, Wolfe et
al. demonstrated a significant correlation between wastewater and case data at
four out of nine sewer sites where more than 10 mpox cases were reported [8].
To
further assess the validity of wastewater surveillance for monitoring mpox
virus DNA and its association with official case numbers, we used an
established and representative wastewater monitoring system for the catchment
area of a Central European city [13].
Material and
methods
Sample collection
and analysis
Wastewater
samples were collected from the local wastewater treatment plant (ProRheno AG) that
receives wastewater from the catchment area of Basel, Switzerland, which has 273,075
inhabitants (including 201,971 in the political district of Canton Basel-City,
Switzerland, 67,388 in the Canton Basel-Country, and 3716 in parts of three
municipalities in Germany and France). Twenty-four-hour composite samples were collected
daily (except for some days, generally on weekends or around the national
holiday on 1 August, when 48- or 72-hour composite samples were collected; table
S1) and contained 500 ml of wastewater. Samples were stored at 4°C for up to 72
hours before further processing. Total nucleic acids were concentrated and
extracted from 40 ml of wastewater using the Maxwell® RSC Enviro Total Nucleic
Acid Kit (Promega) twice weekly. Sampling and RNA extraction were conducted as
part of the wastewater-based surveillance system established for COVID-19. Mpox
virus DNA was detected in the samples collected from 1 July to 28 August 2022 and
in archived samples from 4 August 2021, 11 August 2021, and 20 August 2021 using
the VIASURE mpox Virus Real-Time PCR Detection Kit (Ruwag, Switzerland). The
number of gene copies per litre of wastewater was calculated to account for the
initial wastewater volume (40 ml), the concentration factor during the
pre-extraction procedure (500), and the eluate volume (40 µl); one gene copy
per PCR equals 500 gene copies per litre. The result in cycle threshold (Ct)
values is shown in figure 1. The result was assessed qualitatively as
“positive” (Ct <39) or “negative” (Ct ≥39) (table S1).
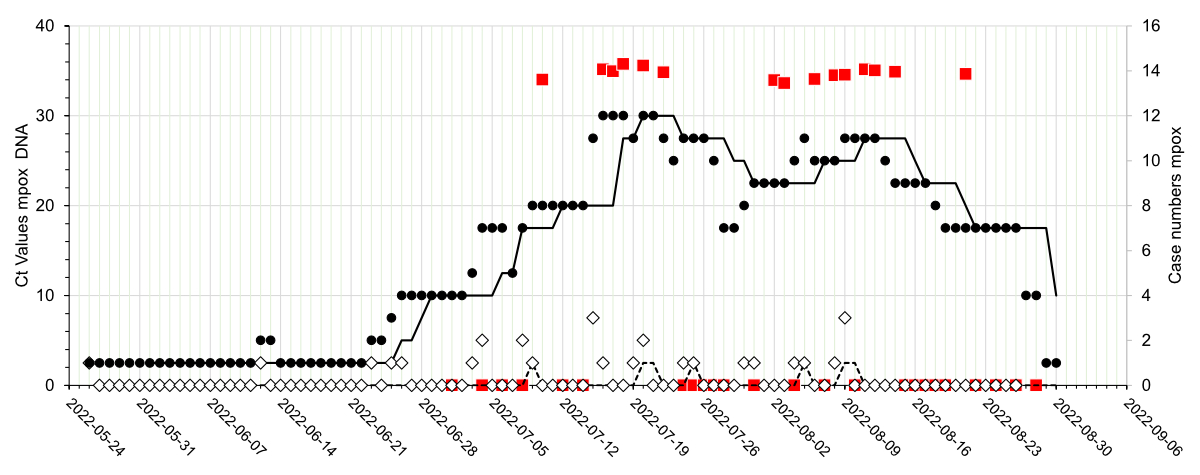
Figure 1Detection of mpox virus DNA in wastewater in relation to mpox (mpox) case numbers
in the catchment area of the city of Basel during the
indicated time period. The mpox virus DNA status of the wastewater samples (n = 39)
is
indicated (red-filled squares; negative samples on the x-axis, positive
samples shown by their Ct values). Open diamonds represent the number of newly
reported mpox cases, and closed circles represent the
number of symptomatic patients. The corresponding seven-day median curves are
shown by black lines (dashed line: newly reported cases, solid line:
symptomatic cases). The results of 48- and 72-hour pooled samples were
attributed to the corresponding later date.
Mpox case
data
Data
on the number of newly diagnosed mpox cases (new cases counted on the day
of diagnosis) and the number of symptomatic cases (accounting for the
symptomatic period of each patient) was captured daily and provided by the
Health Departments of Basel-Stadt and Basel-Landschaft. Symptomatic case data is
based on the mandatory reporting of all PCR- or serological test-confirmed
results as specified by federal law [14] and
on local health authorities’ follow-up calls to all patients. The number of
patients still reporting symptoms was determined each day. A formal study
protocol was not prepared and published in a respective registry.
Statistical
analysis
Logistic
regression analyses with robust standard errors were performed to assess associations
between the detectability of mpox virus DNA in wastewater and daily mpox
cases (absolute numbers and seven-day medians) and the daily number of
symptomatic patients (absolute numbers and seven-day medians). We defined the
outcome as the detectability of mpox virus DNA in wastewater (categorised
as detectable or undetectable). Therefore, odds ratios (ORs) represent the odds
of detecting mpox virus DNA in wastewater per symptomatic case (or number
of daily cases) or per unit of the seven-day median of symptomatic cases (or seven-day
median of the number of daily cases). Furthermore, the number of daily mpox
cases and symptomatic mpox cases on days with positive wastewater samples
was compared to the number of daily cases on days with negative wastewater
samples using the Mann-Whitney U test. All statistical analyses were performed
using STATA 16.0 (Stata Corp., College Station, Texas, USA).
Ethical statement
Because
this study did not involve human research, no ethical consent was required.
Results
Detection of mpox
virus DNA in wastewater
Thirty-nine
wastewater samples were analysed during the 59-day study period (table S1). The
detected Ct values were between 34 and 36 (1–3 gene copies per 20 µl reaction,
equalling 500–1500 gene copies per litre of wastewater). The number of gene
copies detected per litre of wastewater was below the limit of quantification
(10 gene copies/reaction). Therefore, the result was only assessed
qualitatively as positive or negative. Mpox virus DNA was detected in 15
of the 39 samples analysed (table S1) from 10 July until 21 August 2022 (figure
1). The three samples from the 2021 control period were negative for mpox virus
DNA.
Association
between mpox virus DNA detected in wastewater and official case numbers
In
the logistic regression analyses, the positivity of the wastewater samples was significantly
associated with the number of symptomatic cases (OR = 2.18, 95% confidence
interval [CI] = 1.38–3.43, p = 0.001) (figure 2) and with the seven-day median
of symptomatic cases (OR = 1.54, 95% CI = 1.03–2.30, p = 0.035). However,
it was not associated with the number of daily cases (OR = 1.23, 95% CI = 0.53–2.90,
p = 0.629) or their seven-day median (OR = 1.69, 95% CI = 0.21–13.87, p = 0.624).
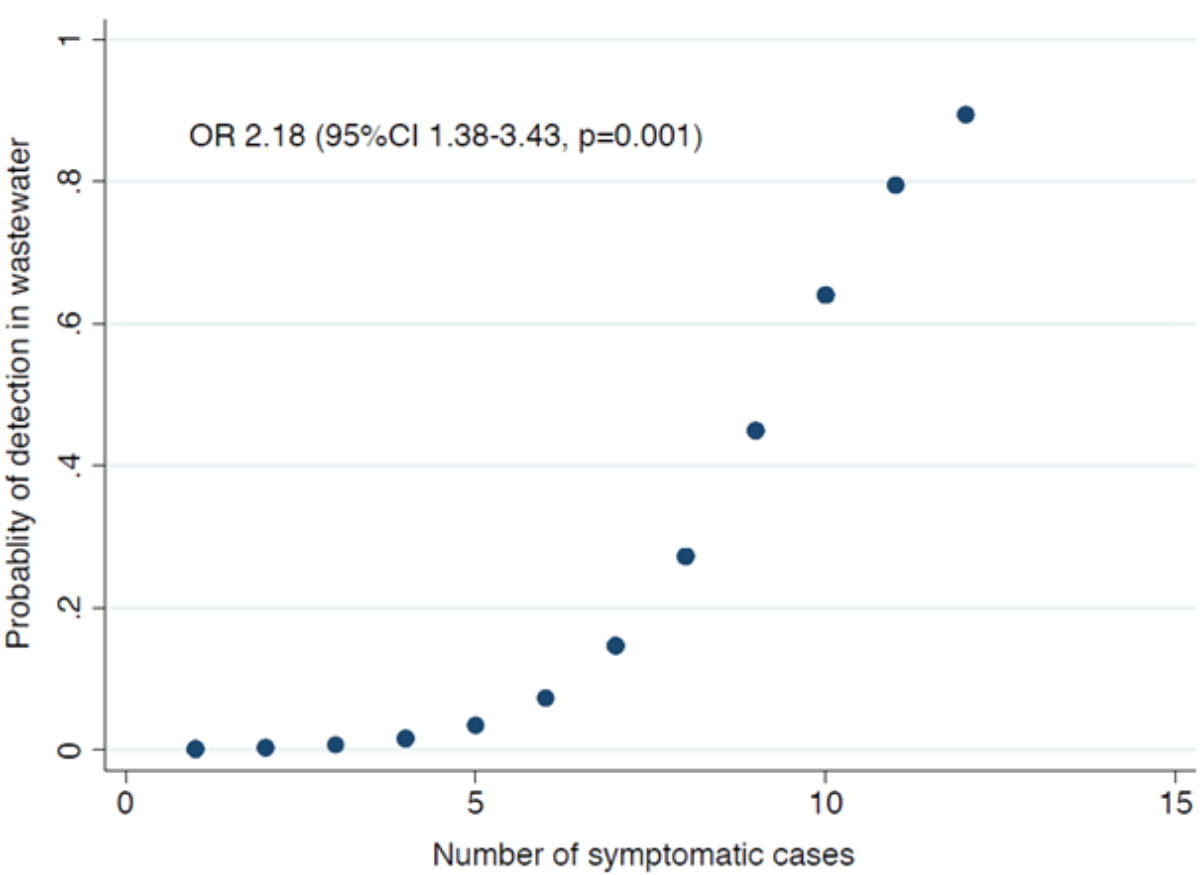
Figure 2Association between the probability of detecting mpox virus DNA in wastewater (sample
positivity) and the number of
symptomatic mpox cases based on logistic regression
analyses. The significance level is indicated.
The number of daily symptomatic cases
differed significantly between days with (median = 11, range = 7–12, interquartile
range [IQR] = 9–12) and days without (median = 8, range = 4–11, IQR = 7–10) mpox
virus DNA detected in wastewater (p = 0.0024) (figure 3). However, newly reported case numbers
did not
differ significantly between days with (median = 0, range = 0–3, IQR = 0–1)
and days without (median = 0, range = 0–2, IQR = 0–1) mpox virus DNA detected
in wastewater (p = 0.877). Therefore, our results suggest an approximate median of
11
reported symptomatic cases (range = 7–12) may be needed to detect mpox
virus DNA in wastewater from a catchment area of approximately 270,000 inhabitants.
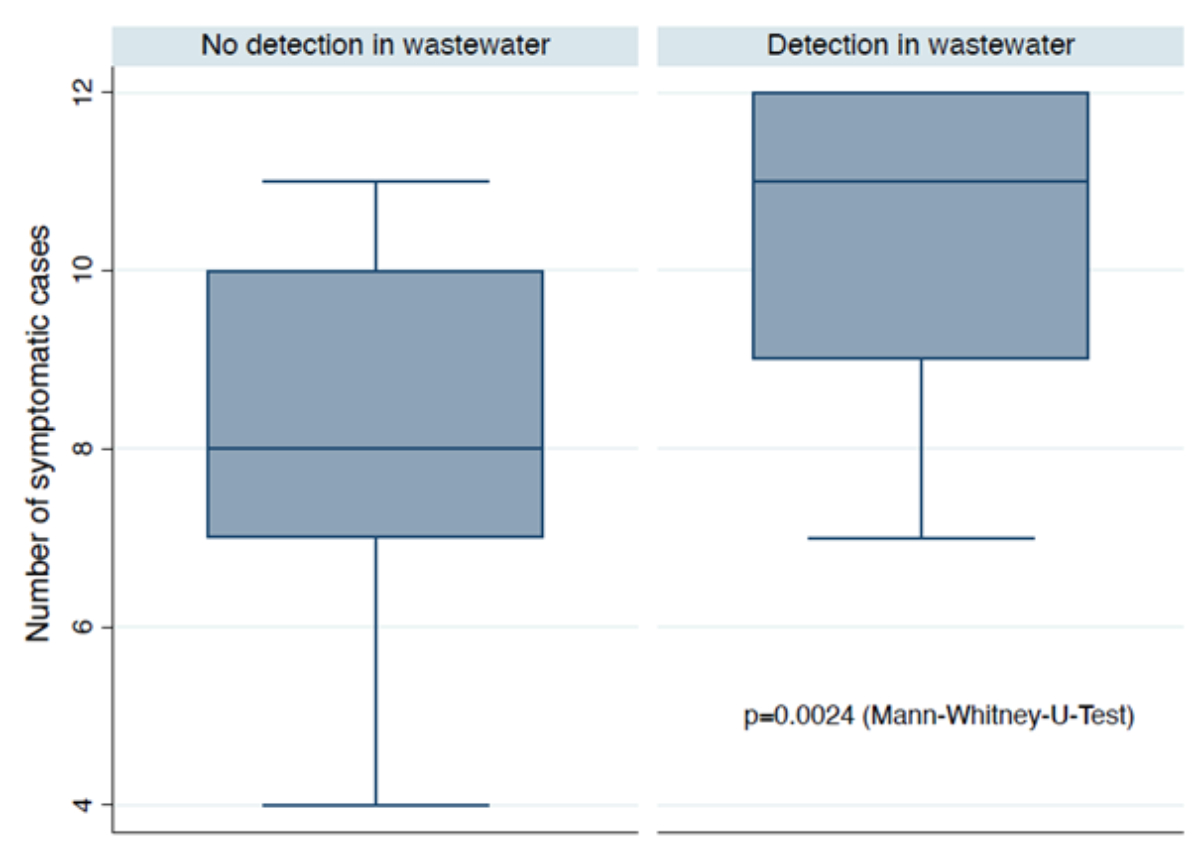
Figure 3Box plots of the number of symptomatic mpox cases on days with and without
mpox virus DNA detected in wastewater.
Discussion
Using
a wastewater-based surveillance system established for COVID-19, mpox
virus DNA was detected in low copy numbers in 15 wastewater samples of a Central
European city from 10 July to 21 August 2022. During this period, there were 0–3
newly reported mpox cases corresponding to 6–12 symptomatic patients. These
numbers might be higher due to underreporting. No data on the actual shedding
of mpox virus into wastewater has been published to date, which would
allow for approximating the rate of underreporting. A comparison of the viral
load in biological samples from 12 patients revealed mpox virus detection
in all saliva and skin lesion samples and most semen, urine, and faeces samples
[15]. Particularly low Ct values
indicating high viral load were obtained for saliva, semen, faeces and skin
lesions. A recent model-based theoretical evaluation by Chen and Bibby
demonstrated that saliva, faeces, and urine contribute the most to the
detectability of mpox virus in wastewater [16].
A
daily shedding load per infected individual of 6 × 107 mpox
virus genome copies was applied, which is at the lower end of the range assumed
for SARS-CoV-2 [17]. Indeed, Wurtzer et
al. suggested that wastewater loads of SARS-CoV-2 and mpox virus were within
the same order of magnitude [10], which
would match our data from a low COVID-19 incidence period (detection limit of
10–20 individuals infected with COVID-19 [13])
and this study (12 individuals with symptomatic mpox). Therefore, mpox
virus DNA is detectable in wastewater, even when officially reported case
numbers are low. Furthermore, our data confirm a high association between the detectability
of mpox virus DNA in wastewater and the number of virus-shedding
symptomatic cases but not with the number of newly diagnosed cases.
Our study was limited by its conduction in a single
city with a limited population size and a limited number of diagnosed and
reported mpox cases during the study period. Nevertheless, our findings
support the utility of wastewater surveillance for detecting emerging
infectious diseases, such as mpox. They should generalise to other
settings with similar population sizes and catchment areas of associated
wastewater treatment plants. Furthermore, while we cannot discount the possibility
of
false positive results, our negative findings for samples from a historic
control period make them unlikely.
Conclusion
Mpox
virus DNA is detectable in wastewater, even when officially reported case
numbers are low. Detectability in wastewater is significantly associated with
the number of symptomatic patients within a catchment area. These findings
support the value of wastewater-based surveillance systems in assessing the prevalence
of emerging and circulating infectious diseases.
Acknowledgments
We are very grateful to the staff at ProRheno AG for their continuous
support. We also thank Christian Fischer and Sam Erny of the Health Department
of the Canton of Basel-Landschaft for helping with the epidemiological data.
Claudia
Bagutti
Kantonales
Laboratorium Basel-Stadt
Kannenfeldstrasse
2
CH-4056
Basel
claudia.bagutti[at]bs.ch
References
1. European Centre for Disease Prevention and Control. Risk assessment: monkeypox multi-country
outbreak. Stockholm: ECDC; 2022 23 May 2022. Available from: https://www.ecdc.europa.eu/en/publications-data/risk-assessment-monkeypox-multi-country-outbreak Last accessed 2023-07-29
2. Isidro J, Borges V, Pinto M, Sobral D, Santos JD, Nunes A, et al. Phylogenomic characterization
and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus.
Nat Med. 2022 Aug;28(8):1569–72. [cited 2023 Jul 29] 10.1038/s41591-022-01907-y
3. Isidro J, Borges V, Pinto M, Sobral D, Santos JD, Nunes A, et al. Addendum: phylogenomic
characterization and signs of microevolution in the 2022 multi-country outbreak of
monkeypox virus. Nat Med. 2022 Oct;28(10):2220–1. [cited 2023 Jul 29] 10.1038/s41591-022-02036-2
4. Taylor L. Monkeypox: WHO to rename disease to prevent stigma. BMJ. 2022 Jun;377:o1489.
[cited 2023 Jul 29] 10.1136/bmj.o1489
5. De Baetselier I, Van Dijck C, Kenyon C, Coppens J, Michiels J, de Block T, et al.;
ITM Monkeypox study group. Retrospective detection of asymptomatic monkeypox virus
infections among male sexual health clinic attendees in Belgium. Nat Med. 2022 Nov;28(11):2288–92.
[cited 2023 Jul 29] 10.1038/s41591-022-02004-w
6. Miura F, van Ewijk CE, Backer JA, Xiridou M, Franz E, Op de Coul E, et al. Estimated
incubation period for monkeypox cases confirmed in the Netherlands, May 2022. Euro
Surveill. 2022 Jun;27(24):2200448. [cited 2023 Jul 29] 10.2807/1560-7917.ES.2022.27.24.2200448
7. Girón-Guzmán I, Díaz-Reolid A, Truchado P, Carcereny A, García-Pedemonte D, Hernáez B,
et al. Spanish wastewater reveals the current spread of Monkeypox virus. Water Res.
2023 Mar;231:119621. [cited 2023 Jul 29] 10.1016/j.watres.2023.119621
8. Wolfe MK, Yu AT, Duong D, Rane MS, Hughes B, Chan-Herur V, et al. Use of Wastewater
for Mpox Outbreak Surveillance in California. N Engl J Med. 2023 Feb;388(6):570–2.
[cited 2023 Jul 29] 10.1056/NEJMc2213882
9. de Jonge EF, Peterse CM, Koelewijn JM, van der Drift AR, van der Beek RF, Nagelkerke E,
et al. The detection of monkeypox virus DNA in wastewater samples in the Netherlands.
Sci Total Environ. 2022 Dec;852:158265. [cited 2023 Jul 29] 10.1016/j.scitotenv.2022.158265
10. Wurtzer S, Levert M, Dhenain E, Boni M, Tournier JN, Londinsky N, et al. First detection
of Monkeypox virus genome in sewersheds in France. Environ Sci Technol Lett. 2022;9(11):991–6.
[cited 2023 Jul 29] 10.1021/acs.estlett.2c00693
11. La Rosa G, Mancini P, Veneri C, Ferraro GB, Lucentini L, Iaconelli M, et al. Detection
of Monkeypox virus DNA in the wastewater of an airport in Rome, Italy: expanding environmental
surveillance to emerging threats. Emerg Infect Dis. 2023;29(1):193–6. [cited 2023
Jul 29] 10.3201/eid2901.221311
12. Mejia EM, Hizon NA, Dueck CE, Lidder R, Daigle J, Wonitowy Q, et al. Exploration of
wastewater surveillance for Monkeypox virus. medRxiv. 2022:2022.11.10.22282091. 10.1101/2022.11.10.22282091 Last accessed 2023-07-29 10.1101/2022.11.10.22282091
13. Bagutti C, Alt Hug M, Heim P, Maurer Pekerman L, Ilg Hampe E, Hübner P, et al. Wastewater
monitoring of SARS-CoV-2 shows high correlation with COVID-19 case numbers and allowed
early detection of the first confirmed B.1.1.529 infection in Switzerland: results
of an observational surveillance study. Swiss Med Wkly. 2022 Jun;152(152):w30202.
[cited 2023 Jul 29] 10.4414/smw.2022.w30202 10.4414/SMW.2022.w30202
14. Federal Office of Public Health. Reporting systems for infectious diseases requiring
notication. Federal Office of Public Health FOPH Bern, Switzerland [https://www.bag.admin.ch/bag/de/home/krankheiten/infektionskrankheiten-bekaempfen/meldesysteme-infektionskrankheiten/meldepflichtige-ik/meldeformulare.html. Last accessed 2023-07-29
15. Suñer C, Ubals M, Tarín-Vicente EJ, Mendoza A, Alemany A, Hernández-Rodríguez Á, et
al.; Movie Group. Viral dynamics in patients with monkeypox infection: a prospective
cohort study in Spain. Lancet Infect Dis. 2023 Apr;23(4):445–53. [cited 2023 Jul 29]
10.1016/S1473-3099(22)00794-0
16. Chen W, Bibby K. Model-based theoretical evaluation of the feasibility of using wastewater-based
epidemiology to monitor monkeypox [Last accessed]. Environ Sci Technol Lett. 2022;9(9):772–8.
10.1021/acs.estlett.2c00496
17. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological
assessment of hospitalized patients with COVID-2019. Nature. 2020 May;581(7809):465–9.
[cited 2023 Jul 29] 10.1038/s41586-020-2196-x
Appendix: supplementary tables
Table S1Sampling
days in July and August 2022, composite sampling type, sample analysis (n= 39), and
qualitative real-time PCR results for mpox virus DNA.
| July 2022 |
August 2022 |
| Sampling days |
Sample type* |
Sample analysis and result** |
Sampling days |
Sample type |
Sample analysis and result |
| 2022-07-01 |
24 hrs |
Negative |
2022-08-01 |
48 hrs |
Positive |
| 2022-07-02 |
48 hrs |
na |
2022-08-02 |
| 2022-07-03 |
2022-08-03 |
24 hrs |
Positive |
| 2022-07-04 |
24 hrs |
Negative |
2022-08-04 |
24 hrs |
Negative |
| 2022-07-05 |
24 hrs |
na |
2022-08-05 |
48 hrs*** |
Negative |
| 2022-07-06 |
24 hrs |
Negative |
2022-08-07 |
| 2022-07-07 |
24 hrs |
na |
2022-08-06 |
24 hrs |
Positive |
| 2022-07-08 |
24 hrs |
Negative |
2022-08-08 |
24 hrs |
Positive |
| 2022-07-09 |
48 hrs |
Positive |
2022-08-09 |
24 hrs |
Positive |
| 2022-07-10 |
2022-08-10 |
24 hrs |
Negative |
| 2022-07-11 |
24 hrs |
na |
2022-08-11 |
24 hrs |
Positive |
| 2022-07-12 |
24 hrs |
Negative |
2022-08-12 |
24 hrs |
Positive |
| 2022-07-13 |
24 hrs |
na |
2022-08-13 |
48 hrs |
Positive |
| 2022-07-14 |
24 hrs |
Negative |
2022-08-14 |
| 2022-07-15 |
48 hrs |
Positive |
2022-08-15 |
24 hrs |
Negative |
| 2022-07-16 |
2022-08-16 |
24 hrs |
Negative |
| 2022-07-17 |
24 hrs |
Positive |
2022-08-17 |
24 hrs |
Negative |
| 2022-07-18 |
24 hrs |
Positive |
2022-08-18 |
24 hrs |
Negative |
| 2022-07-19 |
24 hrs |
na |
2022-08-19 |
24 hrs |
Negative |
| 2022-07-20 |
24 hrs |
Positive |
2022-08-20 |
48 hrs |
Positive |
| 2022-07-21 |
24 hrs |
na |
2022-08-21 |
| 2022-07-22 |
24 hrs |
Positive |
2022-08-22 |
48 hrs |
Negative |
| 2022-07-23 |
48 hrs |
Negative |
2022-08-23 |
24 hrs |
na |
| 2022-07-24 |
2022-08-24 |
24 hrs |
Negative |
| 2022-07-25 |
24 hrs |
Negative |
2022-08-25 |
24 hrs |
na |
| 2022-07-26 |
24 hrs |
Negative |
2022-08-26 |
24 hrs |
Negative |
| 2022-07-27 |
24 hrs |
Negative |
2022-08-27 |
48 hrs |
Negative |
| 2022-07-28 |
24 hrs |
Negative |
2022-08-28 |
| 2022-07-29 |
72 hrs |
Negative |
|
| 2022-07-30 |
| 2022-07-31 |
Table S2 is available for download as a separate file at https://doi.org/10.57187/s.3706.


