Retrospective data analysis for definition of multidrug resistance in gram-negative bacteria – a consensus proposal
DOI: https://doi.org/10.4414/SMW.2022.w30195
Olivier
Friedlia, Irene
Völlmya, Jacques
Schrenzelb, Stephan
Harbarthc, Andreas
Kronenberga, Swiss Centre for Antibiotic Resistance ANRESIS
aInstitute for Infectious Diseases, University of Bern, Bern, Switzerland
bBacteriology Laboratory, Division of Infectious Diseases, University of Geneva Hospitals and Faculty of Medicine, Geneva, Switzerland
cDivisions of Infectious Diseases and Infection Control, University of Geneva Hospitals and Faculty of Medicine, Geneva, Switzerland
Summary
AIM OF THE STUDY: The main objective of this study was to propose a common definition of multidrug-resistant gram-negative organisms (GN-MDRO), which may be used for epidemiological surveillance and benchmarking.
METHODS: In this retrospective data analysis, we used interpreted qualitative susceptibility data (SIR) from blood culture isolates of different gram-negative microorganisms from the ANRESIS database from 2017–2021. We first analysed testing algorithms used by different Swiss laboratories and investigated cross-resistance patterns within antibiotic groups. Comparing these data with existing international definitions, we developed two different GN-MDRO definitions, an extended one for surveillance purposes (ANRESIS-extended) and a more stringent one for clinical purposes, aimed primarily at the identification of difficult-to-treat GN-MDRO (ANRESIS-restricted). Using these novel algorithms, the rates of invasive GN-MDRO identified in our national dataset were compared with international and national definitions: the European Centre for Disease Prevention and Control (ECDC) definition, the Commission for Hospital Hygiene and Infection (KRINKO) definition and the definition proposed by the University Hospital Zurich.
RESULTS: SIR data of a total of 41,785 Enterobacterales, 2,919 , and 419 spp. isolates were used for the analyses. Five antibiotic categories were used for our MDRO definition: aminoglycosides, piperacillin-tazobactam, third- and fourth-generation cephalosporins, carbapenems and fluoroquinolones. Large differences were found between the testing algorithms of the different laboratories. Cross-resistance analysis within an antibiotic group revealed that the substance most likely to be effective against a particular gram-negative bacterium was not preferentially tested (e.g. amikacin for the aminoglycosides). For all bacterial species tested, the highest rates of multidrug-resistant isolates were found using the ECDC-MDR definition, followed by the ANRESIS-extended definition. The number of MDR-Enterobacterales identified using the ANRESIS-restricted definition (n = 627) was comparable to those identified using the KRINKO (n = 622) and UHZ definitions (n = 437). However, the isolates classified as MDR-Enterobacterales according to the KRINKO, UHZ and ANRESIS-restricted definitions (total n = 870) differed considerably. Only 242 of the isolates (27.8%) were uniformly classified as MDRO according to the KRINKO, UHZ and ANRESIS-restricted definitions. Comparable findings were made for Klebsiella spp. and Pseudomonas aeruginosa.
CONCLUSIONS: The application of different MDRO definitions leads to significant differences in not only MDRO rates but also the isolates that are eventually classified as MDRO. Therefore, defining a nationwide MDRO algorithm is crucial if data are compared between hospitals. The definition of a minimal antibiotic susceptibility testing panel would improve comparability further.
Introduction
The spread of antimicrobial resistance is a serious threat to clinical medicine and global public health [1]. In particular, the emergence of gram-negative multidrug-resistant (MDR) organisms (GN-MDRO) in recent decades is concerning [2,3]. MDR-Pseudomonas aeruginosa, MDR-Acinetobacter baumannii and MDR-Enterobacterales are leading causes of morbidity and mortality worldwide [4−6]. The appropriate definition and classification of GN-MDRO not only affects patient care and related costs but must also satisfy several overlapping needs, from patient care to infection prevention control (IPC) programs to reimbursement issues and epidemiological surveillance. No definition of GN-MDRO is uniformly accepted in Switzerland, and it seems difficult to find one single definition suiting all needs [7].
On the international level, different definitions of GN-MDRO exist. In the majority of studies on MDRO, the definition proposed 10 years ago by the European Centre for Disease Prevention and Control (ECDC) has been applied [8]. However, the definition of Magiorakos presumes extensive testing, including 17 antimicrobial categories, of which isolates must be resistant to at least three to be considered MDROs. Due to lack of weighting in the definition of the ECDC, GN-MDRO not only are very frequent but also very heterogeneous, ranging from isolates resistant to ampicillin, minocycline and cotrimoxazole to isolates resistant to imipenem, amikacin and cefepime. Another definition, focused on hospital hygiene purposes, was developed in 2012 by the Commission for Hospital Hygiene and Infection Prevention (KRINKO) at the Robert Koch Institute [9]. This definition does not consider aminoglycosides at all, including only four antibiotic categories. It defines GN-MDRO as phenotypic resistance towards three or four out of these antibiotic groups (named 3-MRGN and 4-MRGN, where MRGN stands for multidrug-resistant gram-negative). Within Switzerland, different MDR definitions – frequently adapted to local testing strategies – are used in individual institutions. One of the best-described local algorithms was published by Wolfensberger et. al for the University Hospital of Zurich (UHZ) [10]. Although this definition is reasonable and comparable with ours, restriction of the MDR definition to a pre-defined testing strategy makes inter-institutional comparisons impossible.
Considering these aspects, we aimed to propose a common definition of GN-MDRO based on national surveillance data. An ideal definition should consider various aspects such as the resistance mechanisms of different bacterial species, the different testing strategies used by different laboratories and the scope of multidrug resistance. It is based on phenotypic resistance patterns and suited mainly for epidemiological surveillance and inter-institutional benchmarking but could also serve as a starting point to streamline infection control practices.
Materials and methods
Study design and setting
In this retrospective data analysis, we extracted antibiotic susceptibility (AST) results from the database of the Swiss Antibiotic Resistance Centre (ANRESIS) [11]. This database contains all AST results from 33 participating laboratories, representing around 87% of all hospitalised patients in Switzerland. The laboratories are geographically distributed across all Swiss regions and include private, university and general hospital laboratories. All laboratories participate in at least one external quality program and are approved by the Swiss Agency for Therapeutic Products (swissmedic). Resistance test results are interpreted locally according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, www.eucast.org
http://www.eucast.org) or Clinical Laboratory Standards Institute (CSLI, www.clsi.org
http://www.clsi.org). "Susceptibility increased exposure" (I, formerly described as "intermediate") was judged as susceptible.Breakpoint changes due to changes in EUCAST definitions were adjusted retrospectively, where possible. This concerns the resistance data for amikacin, tobramycin, gentamicin and imipenem. An isolate was considered resistant to an antibiotic group if the microorganism was resistant to at least one of the antibiotics in that group.
Laboratories routinely send all AST results from all types of clinical samples to ANRESIS, even if the results are not reported to the clinicians. However, for this analysis, we used AST results only from blood culture isolates because these isolates are tested against a broader spectrum of antibiotics. We included all blood culture isolates submitted to ANRESIS between January 2017 and December 2021. Duplicate entries were removed, and only the first date of occurrence in reinfected patients was recorded.
Statistical analysis
Descriptive statistics were applied. The results were presented as numbers and frequencies, which were compared using the chi-squared test; p <0.05 was considered significant. All analyses were performed using the R software environment (version 4.0.2) or KNIME software (4.3.1).
Testing of MDR definitions
We first analysed the proportion of individual antibiotics tested by different laboratories. If more than three antibiotics per antibiotic group were tested, we restricted the analysis to the three most frequently tested antibiotics of a given antibiotic group. Further, for the analysis of cross-resistance within an antibiotic group, we considered each antibiotic group separately and included data only if all antibiotics in that group were tested and an isolate was resistant to at least one of them. Third, we compared our national dataset with international and national definitions of GN-MDRO: the ECDC definition by Magiorakos [8], the KRINKO definition [9] and the definition proposed by the UHZ [10]. We then developed a national definition for MDR-Enterobacterales, P. aeruginosa and Acinetobacter spp., considering international definitions, different national testing strategies and cross-resistance within antibiotic groups. To meet the different needs, we developed two different definitions: a broader extended one, focusing on epidemiological surveillance (“ANRESIS-extended [-EXT]”) and a more restrictive one focusing more on clinical issues (“ANRESIS-restricted [-RST]”).
MDR definitions
Finally, we applied the two ANRESIS definitions, as well as the ECDC, KRINKO and UZH algorithms to our original dataset. The different definitions are summarised in tables 1–3. KRINKO definitions include two MDR categories, “KRINKO-3MRGN” and “KRINKO-4MRGN”, as described.
Table 1Definition criteria and antibiotic panel for Enterobacterales.
|
Antibiotic category
|
ECDC
1
|
KRINKO
|
UHZ
|
ANRESIS-RST
|
ANRESIS-EXT
|
| Aminoglycosides |
Gentamicin or tobramycin or amikacin or netilmicin |
|
At least two of amikacin, gentamicin, tobramycin |
Amikacin and gentamicin |
At least one of amikacin, gentamicin, tobramycin |
| Antipseudomonal penicillins + BLI |
Ticarcillin-clavulanic acid or piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
| Carbapenems |
Ertapenem or imipenem or meropenem or doripenem |
Imipenem and/or meropenem |
At least two of ertapenem, imipenem, meropenem |
Imipenem and/or meropenem |
At least one of ertapenem, imipenem, meropenem |
| Extended-spectrum cephalosporins; third- and fourth-generation cephalosporins |
Cefotaxime or ceftriaxone or ceftazidime or cefepime |
Cefotaxime and/or ceftazidime |
Ceftriaxone and ceftazidime and cefepime |
Ceftazidime and cefepime |
At least one of ceftriaxone, ceftazidime, cefepime |
| Fluoroquinolones |
Ciprofloxacin |
Ciprofloxacin |
Ciprofloxacin and levofloxacin |
Ciprofloxacin |
Ciprofloxacin and/or levofloxacin |
| Definition of MDR |
Resistant to at least 3 out of 17 categories |
Resistant to at least 3 out of 4 categories |
Resistant to at least 3 out of 5 categories |
Resistant to at least 3 out of 5 categories |
Resistant to at least 3 out of 5 categories |
Table 2Definition criteria and antibiotic panel for Pseudomonas aeruginosa.
|
Antibiotic category
|
ECDC
1
|
KRINKO
|
UHZ
|
ANRESIS-RST
|
ANRESIS-EXT
|
| Aminoglycosides |
Gentamicin or tobramycin or amikacin or netilmicin |
|
At least two of amikacin, gentamicin, tobramycin |
Amikacin and/or tobramycin |
At least one of amikacin, gentamicin, tobramycin |
| Antipseudomonal penicillins + BLI |
Ticarcillin-clavulanic acid or piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
| Carbapenems |
Imipenem or meropenem or doripenem |
Imipenem and meropenem |
Imipenem and meropenem |
Meropenem and/or imipenem |
Imipenem and/or meropenem |
| Extended-spectrum cephalosporins; third- and fourth-generation cephalosporins |
Ceftazidime or cefepime |
Ceftazidime and cefepime |
Ceftazidime and cefepime |
Ceftazidime and/or cefepime |
Ceftazidime and/or cefepime |
| Fluoroquinolones |
Ciprofloxacin or levofloxacin |
Ciprofloxacin |
Ciprofloxacin and levofloxacin |
Ciprofloxacin |
Ciprofloxacin and/or levofloxacin |
| Definition of MDR |
Resistant to at least 3 out of 8 categories |
Resistant to at least 3 out of 4 categories |
Resistant to at least 3 out of 5 categories |
Resistant to at least 3 out of 5 categories |
Resistant to at least 3 out of 5 categories |
Table 3Definition criteria and antibiotic panel for Acinetobacter spp.
|
Antibiotic category
|
ECDC
1
|
KRINKO
|
UHZ
|
ANRESIS-RST
|
ANRESIS-EXT
|
| Aminoglycosides |
Gentamicin or tobramycin or amikacin or netilmicin |
|
At least two of amikacin, gentamicin, tobramycin |
Amikacin and/or tobramycin |
At least one of amikacin, gentamicin, tobramycin |
| Antipseudomonal penicillins + BLI |
Ticarcillin-clavulanic acid or piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
Piperacillin-tazobactam |
| Carbapenems |
Imipenem or meropenem or doripenem |
Imipenem and/or meropenem |
Imipenem and/or meropenem |
Imipenem and/or meropenem |
Imipenem and/or meropenem |
| Extended-spectrum cephalosporins; third- and fourth-generation cephalosporins |
Cefotaxime or ceftriaxone or ceftazidime or cefepime |
Cefotaxime and/or ceftazidime |
Ceftazidime and cefepime |
Ceftazidime and cefepime |
Ceftazidime and/or cefepime |
| Fluoroquinolones |
Ciprofloxacin or levofloxacin |
Ciprofloxacin |
Ciprofloxacin and levofloxacin |
Ciprofloxacin and/or levofloxacin |
Ciprofloxacin and/or levofloxacin |
| Definition of MDR |
Resistant to at least 3 out of 9 categories |
Resistant to at least 3 out of 4 categories |
Resistant to at least 3 out of 5 categories |
Resistant to at least 3 out of 5 categories |
Resistant to at least 3 out of 5 categories |
Results
From January 2017 to December 2021, the AST results of 47,264 Enterobacterales, 3,385 , and 456 spp. blood culture isolates were available in the ANRESIS database. After the removal of duplicates, 41,785, 2,919 and 419 isolates, respectively, were used for further analysis.
Testing strategies
The proportion of Enterobacterales tested against individual antibiotics is depicted in figure 1 (laboratory-specific proportions and proportions for P. aeruginosa and Acinetobacter spp., see Appendix, table S1 and figure S2). The three most frequently tested antibiotics of each antibiotic category used in our MDR definition are highlighted. More than 90% of all isolates were tested for ceftriaxone, ceftazidime, cefepime, piperacillin-tazobactam, imipenem and ciprofloxacin. Within the Enterobacterales, only marginal differences in test frequencies were detectable for the most important species (see Appendix table S1). For P. aeruginosa, the antibiotic panels tested differed only slightly and primarily excluded the two antibiotics ceftriaxone and ertapenem, to which P. aeruginosa is intrinsically resistant (Appendix table S1). In Acinetobacter spp. Isolates, third- and fourth-generation cephalosporins and piperacillin-tazobactam were tested less frequently than in other gram-negative species, with large variation between different laboratories. This variability in testing procedures is most likely due to different strategies in communicating to clinicians the possible inducibility of beta-lactam resistance due to AmpC over-expression (Appendix table S1).
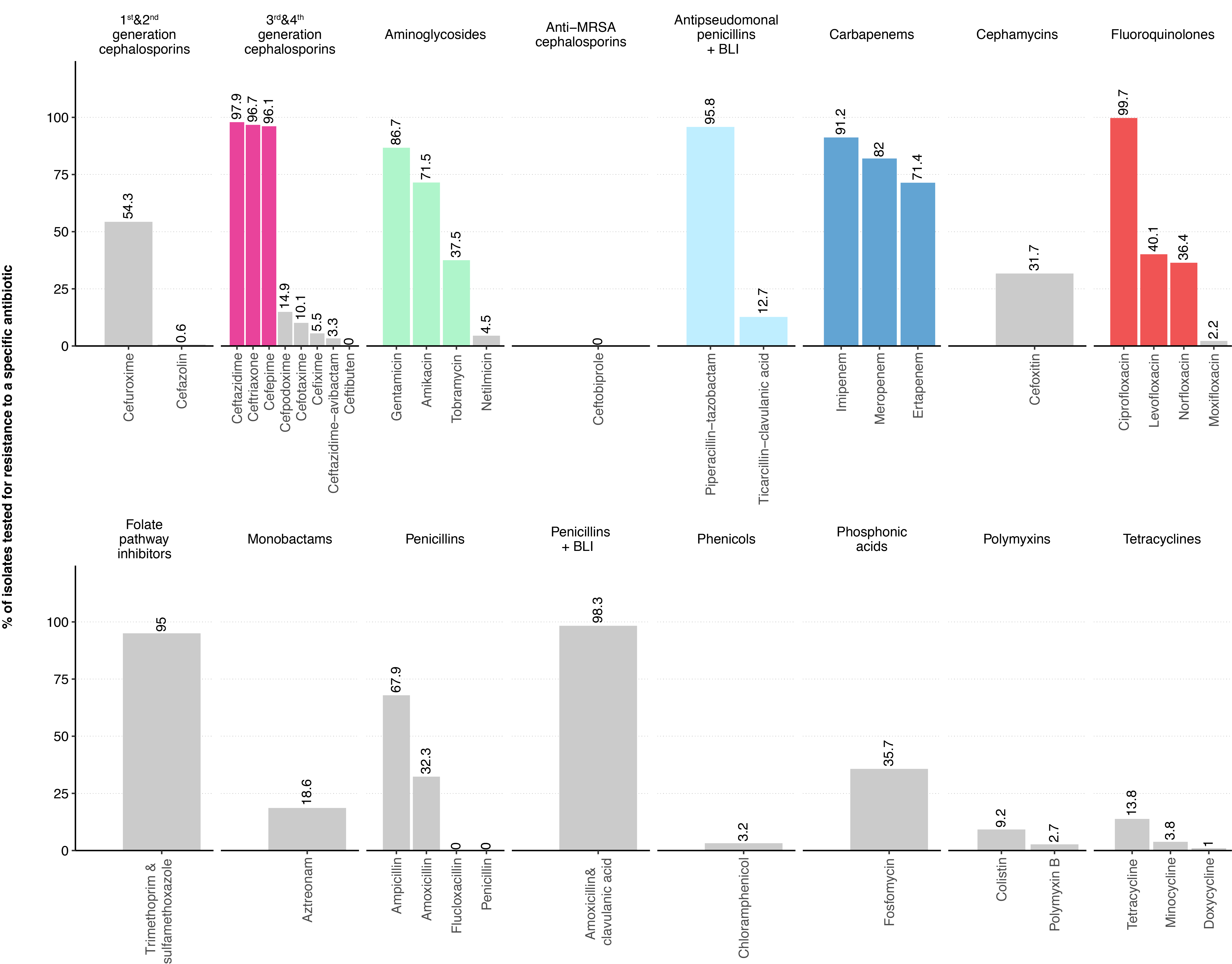
Figure 1 Proportion of Enterobacterales blood culture isolates tested for specific antibiotics among the antibiotic categories defined by the ECDC.
The most frequently tested antibiotics in the five antibiotic categories important for the definition of GN-MDRO are highlighted in colour. BLI, β-lactamase inhibitor; MRSA, methicillin-resistant Staphylococcus aureus.
Cross-resistance
Cross-resistance within antibiotic groups was tested for third- and fourth-generation cephalosporins, carbapenems, aminoglycosides and fluoroquinolones. For this analysis, we only included data if all of the selected (maximum three) antibiotics of a given group were tested and an isolate was resistant to at least one antibiotic. Therefore, we obtained rather low numbers for single antibiotic-microorganism combinations (Appendix table S3). Among Enterobacterales, cefepime was the most active substance among the third- and fourth-generation cephalosporins, while it was amikacin among the aminoglycosides and piperacillin-tazobactam among the antipseudomonal penicillins. Of the carbapenems, isolates were much more frequently resistant to ertapenem than to other antibiotics in this group. Within the fluoroquinolone category, no differences existed in susceptibility to ciprofloxacin or levofloxacin (figure 2, Appendix figure S3). The cross-resistance situation for P. aeruginosa and Acinetobacter spp. isolates was similar.
In contrast to Enterobacterales, spp. isolates were most susceptible to tobramycin among all aminoglycosides. Among the fluoroquinolones, levofloxacin for Acinetobacter spp. and ciprofloxacin for P. aeruginosa were the antibiotics to which the isolates were most likely to still be susceptible.
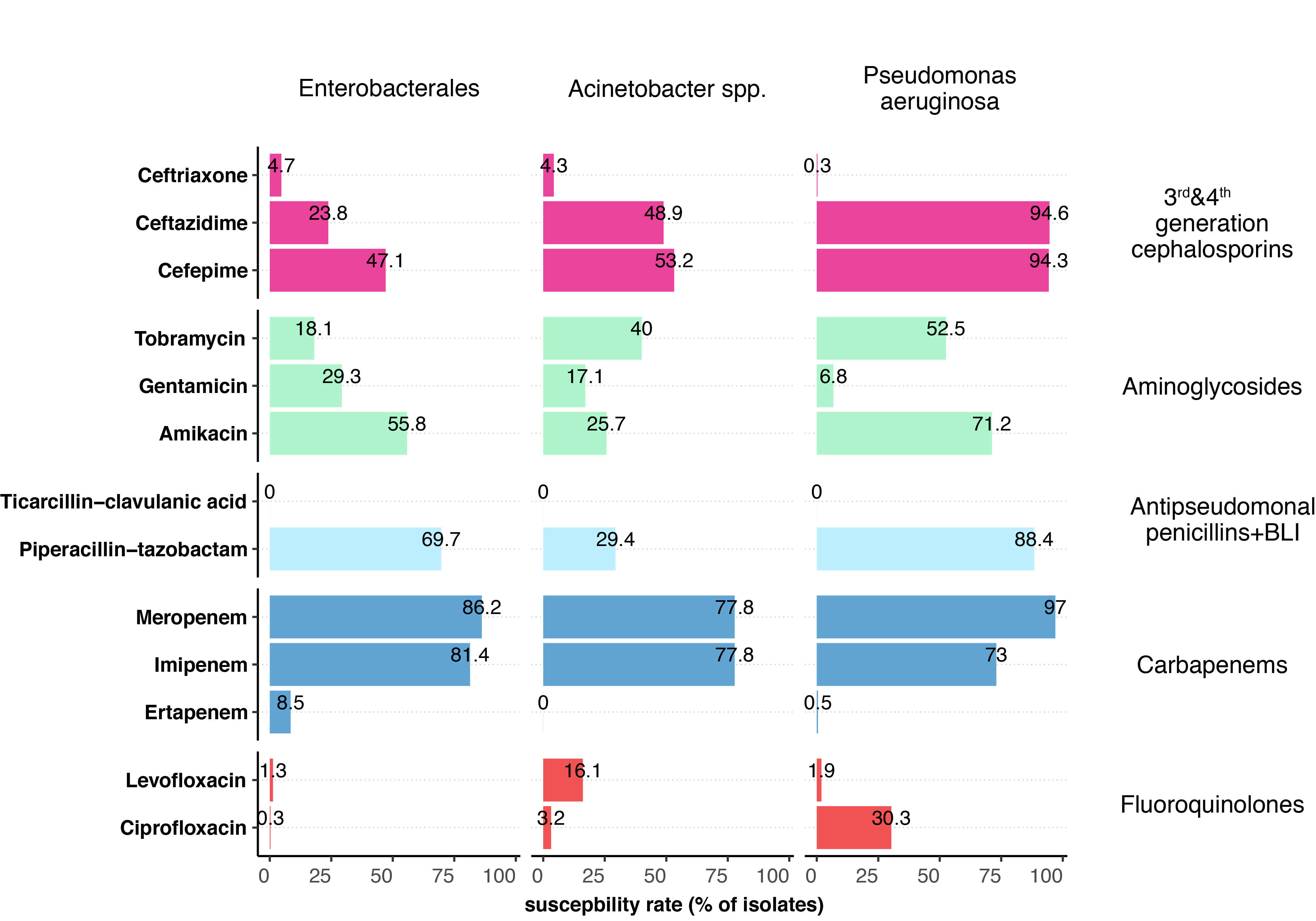
Figure 2 Summary of the cross-resistance situation in the different groups of gram-negative bacteria.
To assess cross-resistance within an antibiotic category, only isolates for which all of the most frequently tested antibiotics of a group were analysed and an isolate was resistant to at least one substance in a group were included in this analysis (for the number of isolates included per antibiotic category, see table S3 in the Appendix). BLI, β-lactamase inhibitor.
Development of MDRO definitions
Based on the test frequencies and the findings on cross-resistance, two different MDRO definitions were established for Enterobacterales (table 1), P. aeruginosa (table 2) and Acinetobacter spp. (table 3): an extended one for early detection of resistance trends (ANRESIS-extended) and one suited more for detection of difficult-to-treat GN-MDRO (ANRESIS-restricted). The antibiotics chosen for the ANRESIS-restricted definition were predominantly those with the highest susceptibility rates within an antibiotic group, whereas for the ANRESIS-extended definition, all frequently tested antibiotics of a group were considered, and resistance was suspected if at least one of the antibiotics was tested resistant. Applying these definitions and the published definitions to our dataset revealed the following numbers of MDRO, summarised in figure 3 (p-values for differences in Appendix table S2).
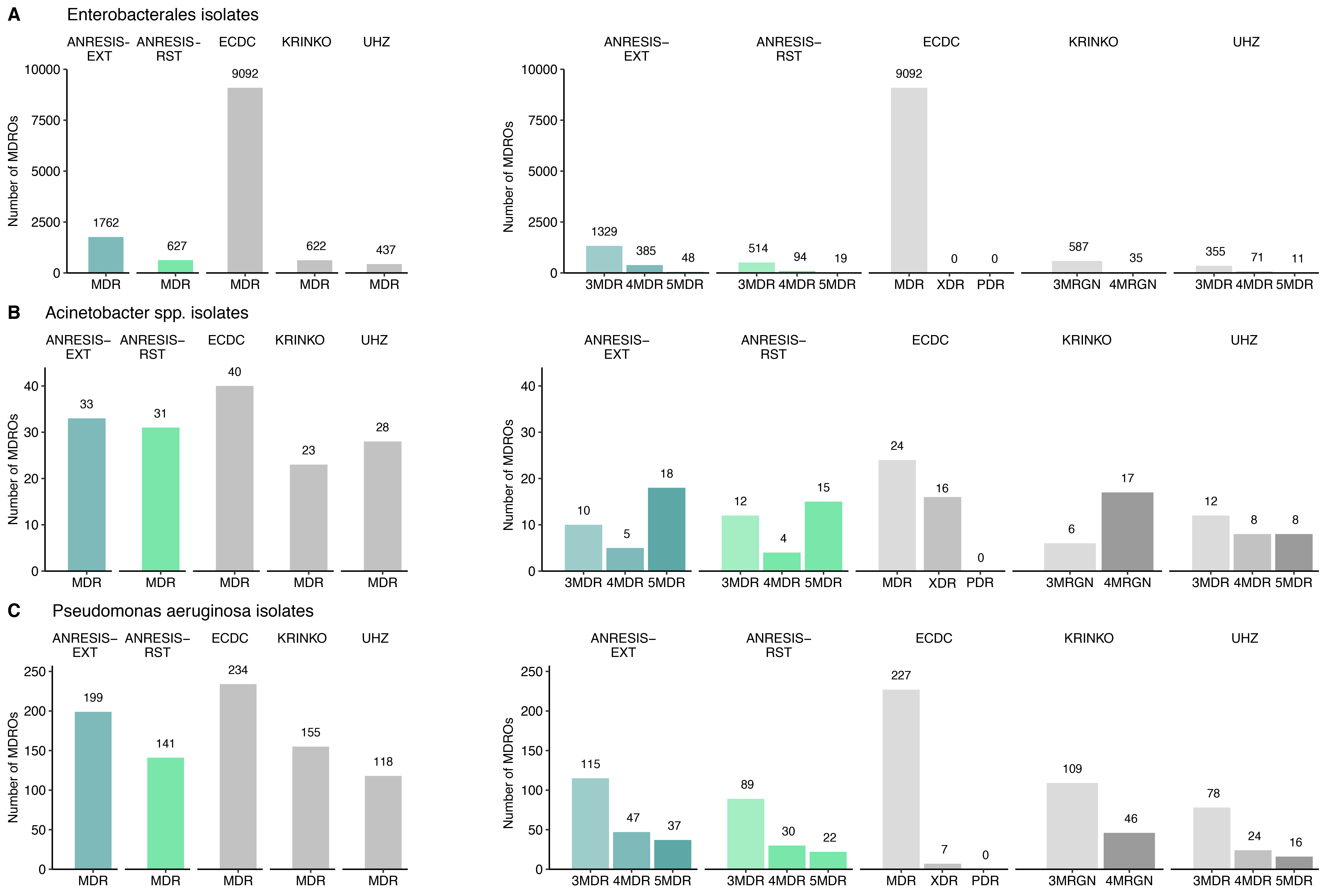
Figure 3 Number of isolates meeting the respective MDRO criteria is displayed.
In the left figures, the total number of isolates (Enterobacterales n = 41,785, Acinetobacter spp. n = 419, Pseudomonas aeruginosa n = 2,919) fulfilling the MDRO criteria is shown. In the right figures, the MDR isolates are displayed according to their degree of multidrug resistance. The number ahead of MDR indicates against how many antibiotic categories an isolate was found to be resistant.
ECDC, European Centre for Disease Prevention and Control; KRINKO, Commission for Hospital Hygiene and Infection Prevention; UHZ, University Hospital Zurich; ANRESIS, Swiss Centre for Antibiotic Resistance; RST, restricted; EXT, extended; MDR, multiple drug resistance; MDRO, multiple drug-resistant organisms; MRGN, multi-resistant gram-negative; PDR, pan drug resistance – isolates are non-susceptible to all agents in all antimicrobial categories; XDR, extensive drug resistance – isolates are non-susceptible to at least one agent in all but two or fewer antimicrobial categories.
Application of different MDRO definitions on the ANRESIS dataset
For all bacterial species tested, the highest rates of multidrug-resistant isolates were found applying the ECDC-MDR definition. The number of MDRO using the ANRESIS-restricted definition was very close to definitions used by KRINKO and the UHZ. The less-stringent ANRESIS-extended definition detected significantly more MDRO in all microorganism groups, most pronounced in Enterobacterales. Although KRINKO, UHZ and ANRESIS-restricted detected comparable numbers of MDRO, isolates identified as MDRO differed essentially between algorithms. Taken together, a total of 870 isolates were classified as MDRO using one of the three definitions. Around a quarter (27.8%) of the isolates were identified as such by all three definitions. The correlation between the MDRO cases found was 70.6% (combined n = 517) with the ANRESIS-restricted and KRINKO definitions and 38.5% (combined n = 296) with the UHZ definition. The correlation between KRINKO and UHZ was 30.1% (combined n = 245; figure 4 A and figure S4 in Appendix). However, all but a single isolate detected by one of these stringent algorithms was included in the less-stringent ANRESIS-extended and ECDC algorithms (figure 4B). For P. aeruginosa and Acinetobacter spp., the overlap was larger, most likely due to more intrinsic resistance and thus more restricted treatment possibilities. Using KRINKO, ANRESIS-restricted and UHZ definitions, 65.2% of identified P. aeruginosa isolates and 67.7% of Acinetobacter spp. isolates overlapped (figures S5 and S6 in the Appendix).
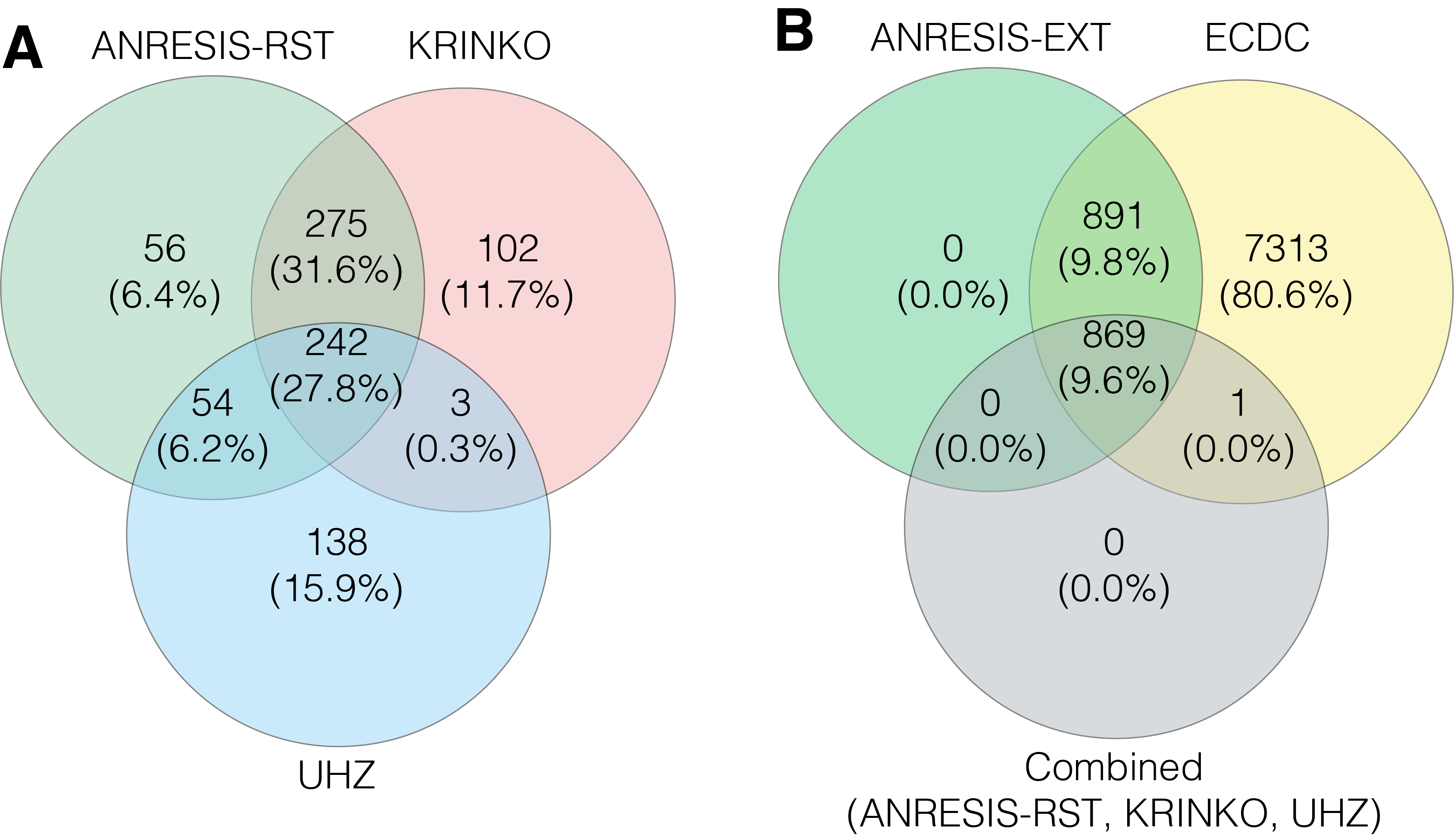
Figure 4 A) Overlap in the number of Enterobacterales isolates classified as MDR following the application of the KRINKO, UHZ and ANRESIS-RST definitions.
B) Overlap in the number of Enterobacterales isolates classified as MDR after application of the ECDC and ANRESIS-EXT definitions and the combined MDR cases found with the KRINKO, UHZ and ANRESIS-RST definitions. MDR, multiple drug resistance; ECDC, European Centre for Disease Prevention and Control; KRINKO, Commission for Hospital Hygiene and Infection Prevention; UHZ, University Hospital Zurich; ANRESIS, Swiss Centre for Antibiotic Resistance; RST, restricted; EXT, extended.
Discussion
Switzerland has no national definition of MDRO yet. The existing MDR definitions considered in our analysis cannot be applied without reservation. The ECDC definition is too general, which means that even relatively harmless microorganisms are classified as MDRO. The KRINKO definition neglects aminoglycosides, whereas the UHZ definition has strict conditions for the antibiotics to be tested so that it cannot be universally applied. Indeed, as the national reference centre of a federal state without national guidelines for antimicrobial resistance (AMR) testing, we must address various testing strategies used in different laboratories. Although nowadays the vast majority of microbiology laboratories use EUCAST guidelines, the selection of antibiotics tested and the reporting of resistance mechanisms depend on local policies. However, continuous surveillance is key to any MDRO control program [3,12], and a common definition is a prerequisite for inter-institutional comparisons and benchmarking.
We therefore selected a bottom-up approach to define MDRO from existing, qualitative, nationwide AMR testing results and compared our approach with international standards. The requirements for this definition were to i) consider the different testing strategies of individual laboratories, ii) allow comparability between different institutes and hospitals within our country, iii) allow comparability with international data, using international definitions, and iv) meet different needs, such as early detection of resistance trends for epidemiologists or detection of difficult-to-treat MDRO for IPC.
We first analysed the antibiotics that are routinely tested. A small number of antibiotics are tested for almost all isolates: cefepime, ceftazidime, ciprofloxacin, piperacillin-tazobactam and imipenem. However, some laboratories were testing these antibiotics rarely or not at all (figure S2, Appendix). Therefore, to increase coverage, we usually included at least two antibiotics per antibiotic group, except for piperacillin-tazobactam, which was tested in nearly all laboratories. The broadest variability in antibiotics tested was observed in the aminoglycoside group.
Second, we analysed cross-resistance within the antibiotic groups. Relevant differences were primarily observed in the third- and fourth-generation cephalosporin group, where cefepime proved the most efficient representative (supported by the literature [13]), and in the aminoglycoside group, where amikacin was the most efficient antibiotic for Enterobacterales, whereas tobramycin was slightly more efficient in Acinetobacter spp. This is also in line with the findings of various studies [14–16]. For the ANRESIS-extended MDRO definition, resistance to at least one antibiotic form a group was sufficient. For the restricted MDRO definition, resistance to the "strongest" antibiotic out of these two groups (cefepime and amikacin) was required. (This had two exceptions: 1. Despite the slightly higher susceptibility rate of tobramycin versus amikacin in Acinetobacter spp., we chose amikacin due to the higher testing frequency for this antibiotic and we additionally aimed for homogenisation with the Enterobacterales definition. 2. Ceftazidime and cefepime were judged as equivalent for P. aeruginosa).
For carbapenems, we consider it reasonable to test either meropenem or imipenem because the test frequency is comparable and cross-resistance is very high. However, for Morganella morganii, Proteus spp. And Providencia spp., either meropenem should be preferred or breakpoints of imipenem adapted accordingly due to intrinsically lower activity of imipenem in these species [17]. Restricting our definition of carbapenem resistance to phenotypic results only may not be sufficient because detection of some carbapenemases as OXA-48 is limited due to weak hydrolysis activity, and additional tests may be needed [18]. Notably, carbapenem-resistant Enterobacterales need additional workup for identification of the resistance genes and should be reported to the Federal Office of Public Health and isolates sent to the National Reference Centre for Early Detection and Surveillance of New Antibiotic Resistance Mechanisms (NARA) in Fribourg [19].
Within the fluoroquinolones, ciprofloxacin is the most frequently tested substance. Ciprofloxacin did not differ from levofloxacin in the cross-resistance analysis, except for Acinetobacter spp. being more susceptible to levofloxacin. Although in our dataset, only 31 Acinetobacter isolates with resistance to fluoroquinolones were included, other studies confirm the higher in vitro activity of newer fluoroquinolones in Acinetobacter spp. [20]. Except for Acinetobacter spp., the inclusion of ciprofloxacin as the only fluoroquinolones used for the ANRESIS-restricted definition seems reasonable.
In summary, for the ANRESIS-restricted definition of MDRO, the following antibiotics should be included in antimicrobial testing panels to achieve full comparability between institutions: amikacin, piperacillin-tazobactam, imipenem or meropenem, cefepime and ciprofloxacin. For the ANRESIS-extended definition, gentamicin and ceftazidime should be added. Although the inclusion of amikacin and cefepime as second drugs of their respective groups is not proposed by the European Antimicrobial Resistance Surveillance Network (EARS-Net), testing of more than one representative of aminoglycosides and third- and fourth-generation cephalosporins is advocated by the Central Asian and European Surveillance of Antimicrobial Resistance network (CAESAR).
Finally, we applied our two ANRESIS algorithms to our dataset and compared the results with other published MDRO definitions. The highest number of MDRO was found by the ECDC definition, in which resistance to three out of a total of up to 17 antibiotic groups (including penicillins) is sufficient for the definition of an MDRO. In this algorithm, in principle, the more antibiotics are tested for an isolate, the higher the probability that an isolate will be classified as ECDC-MDR. As already discussed extensively by KRINKO [21], the algorithm does not weight antibiotic classes in any way. Thus, in our view, it may have its role in pre-defined, longitudinal studies but is suited for neither passive surveillance nor meaningful clinical decisions. On the other hand, the ECDC definition includes antibiotic groups such as glycylcyclines, phenicols and polymyxins rarely routinely tested in Swiss laboratories.
In terms of numbers of detected MDRO, the ECDC algorithm is followed by the ANRESIS-extended algorithm, which seems reasonable because this algorithm includes multiple antibiotics per antibiotic group and is intended as an "early warning system", even detecting MDRO without broad cross-resistance within antibiotic groups. Interestingly, all but one isolate identified with the three algorithms (ANRESIS-restrictive, KRINKO, and UHZ) are included in the ANRESIS-extended output.
The more stringent ANRESIS-restrictive algorithm matched reasonably well with the KRINKO and UHZ definitions in terms of numbers of detected MDRO. Although the number of MDRO cases identified is very similar in these three algorithms, the poor overlap in isolates classified as MDRO is surprising. This illustrates that small differences between two algorithms can lead to considerable differences. (For example, the difference in the MDRO cases identified using the ANRESIS-restricted and UHZ definitions is almost entirely due to the different definition for the aminoglycosides. Although resistance to two out of three aminoglycosides is sufficient in the UHZ algorithm, resistance to amikacin is mandatory in the ANRESIS-restricted algorithm, which was not achieved in 45% of the UHZ-aminoglycoside resistant microorganisms.) The implementation of national testing requirements would improve the consistency and comparability of Swiss data. In addition, the MDRO definition could probably be simplified and ideally even adapted to existing international definitions. We hope that our analysis will catalyse the development of national testing guidelines by the responsible organisations.
The strength of our study lies in its bottom-up strategy, using real-world data from 33 different laboratories, distributed all over Switzerland. So far, MDRO definitions have mostly been provided top-down from single laboratories, which applied their definition to a pre-defined collection of microorganisms, using standardised antibiotic test panels. At least in Switzerland, where the health system is under the sovereignty of the cantons, and stringent national guidelines for AST are missing, different testing strategies must be considered when defining an MDRO algorithm. Having access to a huge dataset, we were able to develop and test different MDRO algorithms and compare them to other definitions.
Our study has several limitations. Firstly, resistance mechanisms should ideally guide MDRO definitions. Because resistance mechanisms usually are not available in routine resistance datasets, our definition (as well as definitions from the literature) relies on phenotypic resistance patterns. However, resistance patterns allow drawing conclusions on underlying resistance mechanisms in most cases. For example, in Switzerland, 93–96% of third-generation cephalosporin–resistant E. coli harbour an extended-spectrum beta-lactamase [22], whereas other rarer Enterobacterales such as Enterobacter spp., Citrobacter freundii, Hafnia alvei, M. morganii, Serratia marcescens and Providencia spp. frequently carry an inducible, chromosome-encoded AmpC: cephalosporinase [23]. Secondly, we do not have any data on treatment outcomes, and the impact of the MDR definition on morbidity and mortality could not be studied. Notably, other studies have shown that MDRO infections are associated with higher mortality [24]. Thirdly, our definition has been developed for surveillance purposes. Modification of the proposed definitions may be needed for different requirements, such as clinical treatment decisions, isolation precaution measures or MDRO reimbursement negotiations with health care insurers. We hope that our detailed analysis will stimulate discussion among other specialists such as microbiologists, infectious diseases specialists and hospital hygiene specialists. Finally, newer and reserve antibiotics such as tigecycline, colistin, ceftolozane-tazobactam and ceftazidime-avibactam are not included in our algorithm. Except for the ECDC definition, they are also not included in other algorithms. Most of these antibiotics are not tested routinely, and resistance in Switzerland remains low.
Conclusions
The application of different MDRO algorithms to a database resulted in very diverse outputs, strongly influenced even by small differences in MDRO definitions, if not in number, then at least in the identification of single MDRO isolates. Therefore, defining a nationwide MDRO algorithm is crucial if data are compared between hospitals. The definition of a minimal antibiotic susceptibility testing panel would improve comparability further.
Olivier Friedli, Ph.D.
Institute for Infectious Diseases, University of Bern
Friedbühlstrasse 51
CH-3010 Bern
olivier.friedli[at]unibe.ch
References
1.
Carlet J
,
Collignon P
,
Goldmann D
,
Goossens H
,
Gyssens IC
,
Harbarth S
, et al.
Society’s failure to protect a precious resource: antibiotics. Lancet. 2011 Jul;378(9788):369–71. https://doi.org/10.1016/S0140-6736(11)60401-7
2.
Martens E
,
Demain AL
. The antibiotic resistance crisis, with a focus on the United States. J Antibiot (Tokyo). 2017 May;70(5):520–6. https://doi.org/10.1038/ja.2017.30
3.
Tacconelli E
,
Sifakis F
,
Harbarth S
,
Schrijver R
,
van Mourik M
,
Voss A
, et al.; EPI-Net COMBACTE-MAGNET Group
. Surveillance for control of antimicrobial resistance. Lancet Infect Dis. 2018 Mar;18(3):e99–106. https://doi.org/10.1016/S1473-3099(17)30485-1
4.
Mehrad B
,
Clark NM
,
Zhanel GG
,
Lynch JP 3rd
. Antimicrobial resistance in hospital-acquired gram-negative bacterial infections. Chest. 2015 May;147(5):1413–21. https://doi.org/10.1378/chest.14-2171
5.
Stewardson AJ
,
Marimuthu K
,
Sengupta S
,
Allignol A
,
El-Bouseary M
,
Carvalho MJ
, et al.
Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis. 2019 Jun;19(6):601–10. https://doi.org/10.1016/S1473-3099(18)30792-8
6.
Vincent JL
,
Rello J
,
Marshall J
,
Silva E
,
Anzueto A
,
Martin CD
, et al.; EPIC II Group of Investigators
. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009 Dec;302(21):2323–9. https://doi.org/10.1001/jama.2009.1754
7.
Roth JA
,
Hornung-Winter C
,
Radicke I
,
Hug BL
,
Biedert M
,
Abshagen C
, et al.
Direct Costs of a Contact Isolation Day: A Prospective Cost Analysis at a Swiss University Hospital. Infect Control Hosp Epidemiol. 2018 Jan;39(1):101–3. https://doi.org/10.1017/ice.2017.258
8.
Magiorakos AP
,
Srinivasan A
,
Carey RB
,
Carmeli Y
,
Falagas ME
,
Giske CG
, et al.
Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012 Mar;18(3):268–81. https://doi.org/10.1111/j.1469-0691.2011.03570.x
9.
KRINKO
. Anforderungen an die Hygiene bei der Aufbereitung von Medizinprodukten. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012 Oct;55(10):1244–310. https://doi.org/10.1007/s00103-012-1548-6
10.
Wolfensberger A
,
Kuster SP
,
Marchesi M
,
Zbinden R
,
Hombach M
. The effect of varying multidrug-resistence (MDR) definitions on rates of MDR gram-negative rods. Antimicrob Resist Infect Control. 2019 Nov;8(1):193. https://doi.org/10.1186/s13756-019-0614-3
11.
Gasser M
,
Schrenzel J
,
Kronenberg A
. für das Schweizerische Zentrum für Antibiotikaresistenzen. Aktuelle Entwicklung der Antibiotikaresistenzen in der Schweiz. Swiss Med Forum. 2018;18(46):943-949. https://doi.org/https://doi.org/10.4414/smf.2018.03404
12.
Tacconelli E
,
Buhl M
,
Humphreys H
,
Malek V
,
Presterl E
,
Rodriguez-Baño J
, et al.; EUCIC StopNegative group
. Analysis of the challenges in implementing guidelines to prevent the spread of multidrug-resistant gram-negatives in Europe. BMJ Open. 2019 May;9(5):e027683. https://doi.org/10.1136/bmjopen-2018-027683
13.
Ramirez MS
,
Tolmasky ME
. Amikacin: Uses, Resistance, and Prospects for Inhibition. Molecules. 2017 Dec;22(12):E2267. https://doi.org/10.3390/molecules22122267
14.
Van Looveren M
,
Goossens H
,
Group AS
; ARPAC Steering Group
. Antimicrobial resistance of Acinetobacter spp. in Europe. Clin Microbiol Infect. 2004 Aug;10(8):684–704. https://doi.org/10.1111/j.1469-0691.2004.00942.x
15.
Nowak J
,
Zander E
,
Stefanik D
,
Higgins PG
,
Roca I
,
Vila J
, et al.; MagicBullet Working Group WP4
. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother. 2017 Dec;72(12):3277–82. https://doi.org/10.1093/jac/dkx322
16.
Ekkelenkamp MB
,
Cantón R
,
Díez-Aguilar M
,
Tunney MM
,
Gilpin DF
,
Bernardini F
, et al.
Susceptibility of Pseudomonas aeruginosa Recovered from Cystic Fibrosis Patients to Murepavadin and 13 Comparator Antibiotics. Antimicrob Agents Chemother. 2020 Jan;64(2):e01541-19. https://doi.org/10.1128/AAC.01541-19
17.
The European Committee on Antimicrobial Susceptibility
. 2021, Testing.https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Expert_Rules/2021/Intrinsic_Resistance_and_Unusual_Phenotypes_Tables_v3.3_20211018.pdf
18.
Lutgring JD
,
Limbago BM
. The Problem of Carbapenemase-Producing-Carbapenem-Resistant-Enterobacteriaceae Detection. J Clin Microbiol. 2016 Mar;54(3):529–34. https://doi.org/10.1128/JCM.02771-15
19.
NARA
, The National Reference Center for Emerging Antibiotic Resistance, unifr.ch/med/nara/ (Accessed 11 Jan. 2022).
20.
Heinemann B
,
Wisplinghoff H
,
Edmond M
,
Seifert H
. Comparative activities of ciprofloxacin, clinafloxacin, gatifloxacin, gemifloxacin, levofloxacin, moxifloxacin, and trovafloxacin against epidemiologically defined Acinetobacter baumannii strains. Antimicrob Agents Chemother. 2000 Aug;44(8):2211–3. https://doi.org/10.1128/AAC.44.8.2211-2213.2000
21.
Kaase M
. MRGN: neue Klassifikation für multiresistente gramnegative Bakterien. Laboratoriumsmedizin (Berl). 2013;37(6):299–304. https://doi.org/10.1515/labmed-2012-0067
22.
Kronenberg A
,
Hilty M
,
Endimiani A
,
Muhlemann K
. Temporal trends of extended-spectrum cephalosporin-resistant Escherichia coli and Klebsiella pneumoniae isolates in in- and outpatients in Switzerland, 2004 to 2011. Euro Surveill. 2013 May;18(21):20484. Available from: https://www.ncbi.nlm.nih.gov/pubmed/23725981 https://doi.org/10.2807/ese.18.21.20484-en
23.
Jacoby, G. A.
(2009, Jan). AmpC beta-lactamases. Clin Microbiol Rev, 22(1), 161-182, Table of Contents. https://doi.org/https://doi.org/10.1128/CMR.00036-08
24.
Huh K
,
Chung DR
,
Ha YE
,
Ko JH
,
Kim SH
,
Kim MJ
, et al.; Korean Antimicrobial Resistance Surveillance Network (KARS-Net) Investigators
. Impact of Difficult-to-Treat Resistance in Gram-negative Bacteremia on Mortality: Retrospective Analysis of Nationwide Surveillance Data. Clin Infect Dis. 2020 Dec;71(9):e487–96. https://doi.org/10.1093/cid/ciaa084
Appendix
Proportion of blood culture isolates tested for certain antibiotics
Table S1Percentage of isolates that were tested for specific antibiotic substances.
|
Antibiotic group
|
Substance
|
ENT
|
ENBT
|
ESCH
|
KLEB
|
ANBT
|
PSMN
|
| Total number of isolates |
|
41,785 |
2,089 |
30,096 |
8,626 |
419 |
2,919 |
| First- and second- generation cephalosporins |
Cefazolin |
0.6 |
0.6 |
0.6 |
0.5 |
1.2 |
0.5 |
| Cefuroxime |
54.3 |
43.9 |
55.7 |
52.2 |
20.5 |
20.2 |
| Third- and fourth- generation cephalosporins |
Cefepime |
96.1 |
94.7 |
96.3 |
96.6 |
49.9 |
97.1 |
| Cefixime |
5.5 |
4.8 |
5.4 |
5.2 |
0.2 |
|
| Cefotaxime |
10.1 |
10.1 |
9.7 |
10.2 |
0.7 |
4.1 |
| Cefpodoxime |
14.9 |
17.2 |
14.4 |
15.6 |
4.8 |
7.2 |
| Ceftazidime |
97.9 |
94.2 |
98.4 |
98.2 |
51.5 |
94.8 |
| Ceftazidime-avibactam |
3.3 |
3.3 |
3.3 |
3.9 |
4.6 |
8.5 |
| Ceftriaxone |
96.7 |
92.6 |
97.1 |
96.8 |
26.7 |
28.4 |
| Aminoglycosides |
Amikacin |
71.5 |
76.5 |
71.1 |
73.9 |
79.8 |
87.1 |
| Gentamicin |
86.7 |
86.3 |
87.1 |
86.2 |
83.7 |
67.9 |
| Netilmicin |
4.5 |
5.7 |
4.4 |
4.8 |
8.3 |
7.2 |
| Tobramycin |
37.5 |
40.5 |
37.1 |
38.2 |
72.0 |
78.2 |
| Anti-MRSA cephalosporins |
Ceftobiprole |
0.0 |
|
0.0 |
0.1 |
0.5 |
0.2 |
| Antipseudomonal penicillins + BLI |
Piperacillin-tazobactam |
95.8 |
96.5 |
96.0 |
95.9 |
49.9 |
96.2 |
| Ticarcillin-clavulanic acid |
12.7 |
13.4 |
12.5 |
12.8 |
10.3 |
14.2 |
| Carbapenems |
Ertapenem |
71.4 |
67.7 |
71.9 |
71.2 |
7.8 |
15.8 |
| Imipenem |
91.2 |
91.1 |
91.7 |
91.7 |
92.2 |
93.6 |
| Meropenem |
82.0 |
82.4 |
81.6 |
83.9 |
85.8 |
85.1 |
| Cephamycins |
Cefoxitin |
31.7 |
34.8 |
31.0 |
34.5 |
8.1 |
11.7 |
| Fluoroquinolones |
Ciprofloxacin |
99.7 |
99.6 |
99.7 |
99.8 |
95.9 |
99.0 |
| Levofloxacin |
40.1 |
42.3 |
39.2 |
42.3 |
60.9 |
60.9 |
| Moxifloxacin |
2.2 |
2.0 |
2.4 |
1.8 |
|
0.0 |
| Norfloxacin |
36.4 |
37.6 |
36.0 |
37.3 |
4.4 |
9.1 |
| Folate pathway inhibitors |
Co-trimoxazole |
95.0 |
95.4 |
95.1 |
95.3 |
90.3 |
46.8 |
| Monobactams |
Aztreonam |
18.6 |
19.3 |
18.4 |
19.7 |
14.7 |
32.5 |
| Penicillins |
Amoxicillin |
32.3 |
30.6 |
32.7 |
32.4 |
3.5 |
3.7 |
| Ampicillin |
67.9 |
64.5 |
69.1 |
64.3 |
25.1 |
25.3 |
| Penicillins + BLI |
Co-amoxiclav |
98.3 |
94.5 |
98.9 |
98.3 |
30.8 |
30.5 |
| Phenicols |
Chloramphenicol |
3.2 |
3.4 |
3.1 |
3.4 |
4.8 |
3.6 |
| Phosphonic acids |
Fosfomycin |
35.7 |
31.9 |
36.5 |
34.3 |
7.1 |
7.4 |
| Polymyxins |
Colistin |
9.2 |
9.0 |
8.7 |
9.9 |
13.6 |
19.7 |
| Polymyxin B |
2.7 |
3.0 |
2.7 |
2.5 |
2.8 |
2.8 |
| Tetracyclines |
Doxycycline |
1.0 |
0.7 |
0.9 |
1.0 |
|
|
| Minocycline |
3.8 |
5.1 |
3.3 |
4.2 |
1.2 |
7.0 |
| Tetracycline |
13.8 |
14.8 |
13.1 |
14.3 |
6.2 |
6.6 |
Table S2P-values from pairwise comparisons of the number of MDR isolates from Figure 3 using the chi-squared test.
| Enterobacterales |
|
ANRESIS-EXT |
ANRESIS-RST |
ECDC |
KRINKO |
UHZ |
| ANRESIS-EXT |
|
<0.001 |
<0.001 |
<0.001 |
<0.001 |
| ANRESIS-RST |
|
|
<0.001 |
0.909 |
<0.001 |
| ECDC |
|
|
|
<0.001 |
<0.001 |
| KRINKO |
|
|
|
|
<0.001 |
| UHZ |
|
|
|
|
|
|
|
|
|
|
|
|
Acinetobacter spp. |
|
ANRESIS-EXT |
ANRESIS-RST |
ECDC |
KRINKO |
UHZ |
| ANRESIS-EXT |
|
0.897 |
0.462 |
0.213 |
0.595 |
| ANRESIS-RST |
|
|
0.321 |
0.325 |
0.787 |
| ECDC |
|
|
|
0.036 |
0.164 |
| KRINKO |
|
|
|
|
0.563 |
| UHZ |
|
|
|
|
|
|
|
|
|
|
|
|
Pseudomonas aeruginosa
|
|
ANRESIS-EXT |
ANRESIS-RST |
ECDC |
KRINKO |
UHZ |
| ANRESIS-EXT |
|
0.001 |
0.089 |
0.018 |
<0.001 |
| ANRESIS-RST |
|
|
<0.001 |
0.438 |
0.162 |
| ECDC |
|
|
|
<0.001 |
<0.001 |
| KRINKO |
|
|
|
|
0.026 |
| UHZ |
|
|
|
|
|
Table S3Number of isolates considered for the cross-resistance analysis.
|
Microorganism
|
Antibiotic category
|
n*
|
n
total
**
|
| Enterobacterales |
Third- and fourth-generation cephalosporins |
3,875 |
41,785 |
| Aminoglycosides |
916 |
41,785 |
| Antipseudomonal penicillins + BLI |
887 |
41,785 |
| Carbapenems |
188 |
41,785 |
| Fluoroquinolones |
2,143 |
41,785 |
|
Acinetobacter spp. |
Third- and fourth-generation cephalosporins |
47 |
419 |
| Aminoglycosides |
35 |
419 |
| Antipseudomonal penicillins + BLI |
17 |
419 |
| Carbapenems |
27 |
419 |
| Fluoroquinolones |
31 |
419 |
|
Pseudomonas aeruginosa
|
Third- and fourth-generation cephalosporins |
662 |
2,919 |
| Aminoglycosides |
59 |
2,919 |
| Antipseudomonal penicillins + BLI |
121 |
2,919 |
| Carbapenems |
434 |
2,919 |
| Fluoroquinolones |
208 |
2,919 |
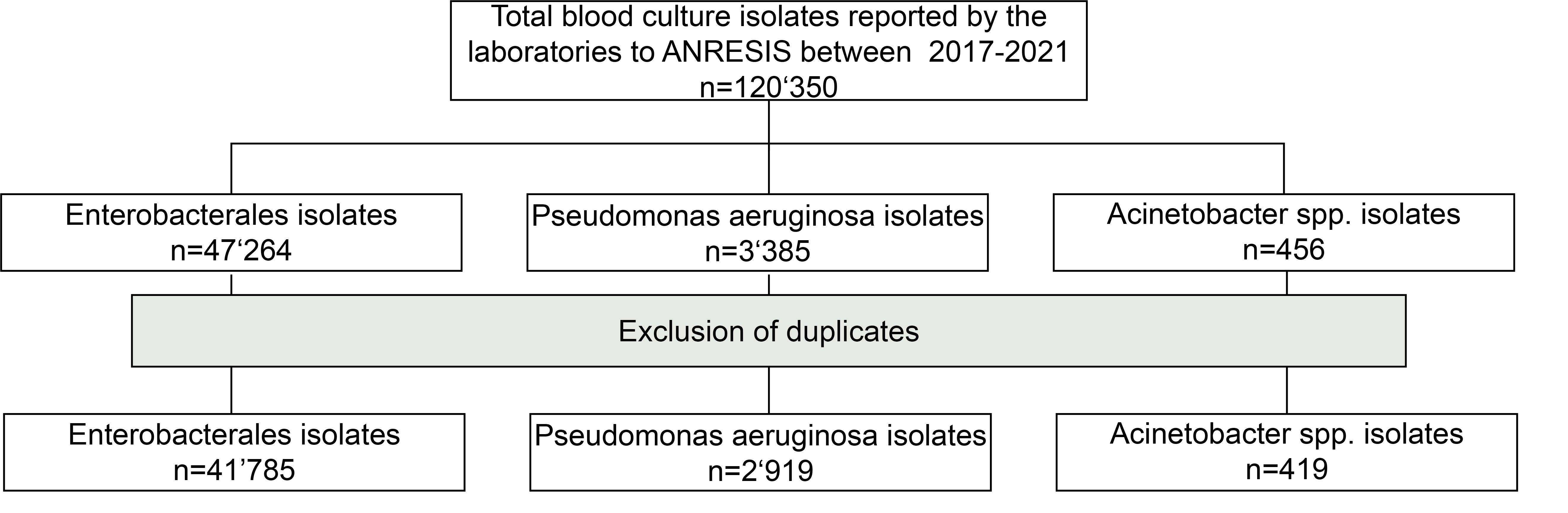
Figure S1 Flowchart of the selection process of isolates submitted to ANRESIS between 2017 and 2021.
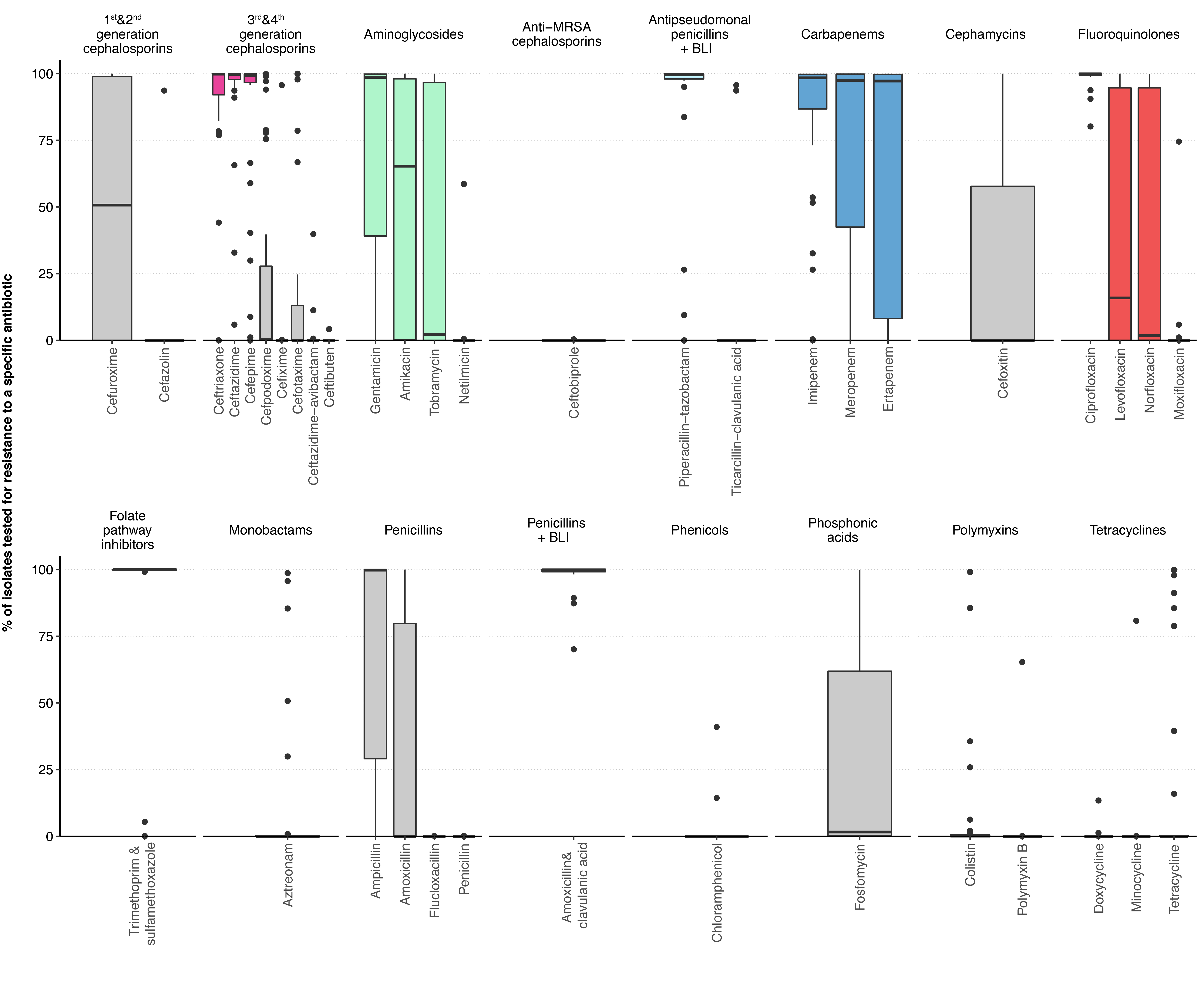
Figure S2 Proportion of Enterobacterales blood culture isolates per laboratory tested for certain antibiotics among the antibiotic categories defined by the ECDC.
The most frequently tested antibiotics in the five antibiotic categories important for the definition of gram-negative MDR are highlighted in colour. Boxplot with horizontal line for the median of the proportion of isolates tested for an antibiotic per laboratory. Boxes represent the first and third quartile, whiskers extend either to the extreme values of the observations or to 1.5 times the interquartile range, and the dots indicate the laboratories declared as outliers. BLI, β-lactamase inhibitor; ECDC, European Centre for Disease Prevention and Control; MDR, multiple drug resistance; MRSA, methicillin-resistant Staphylococcus aureus.
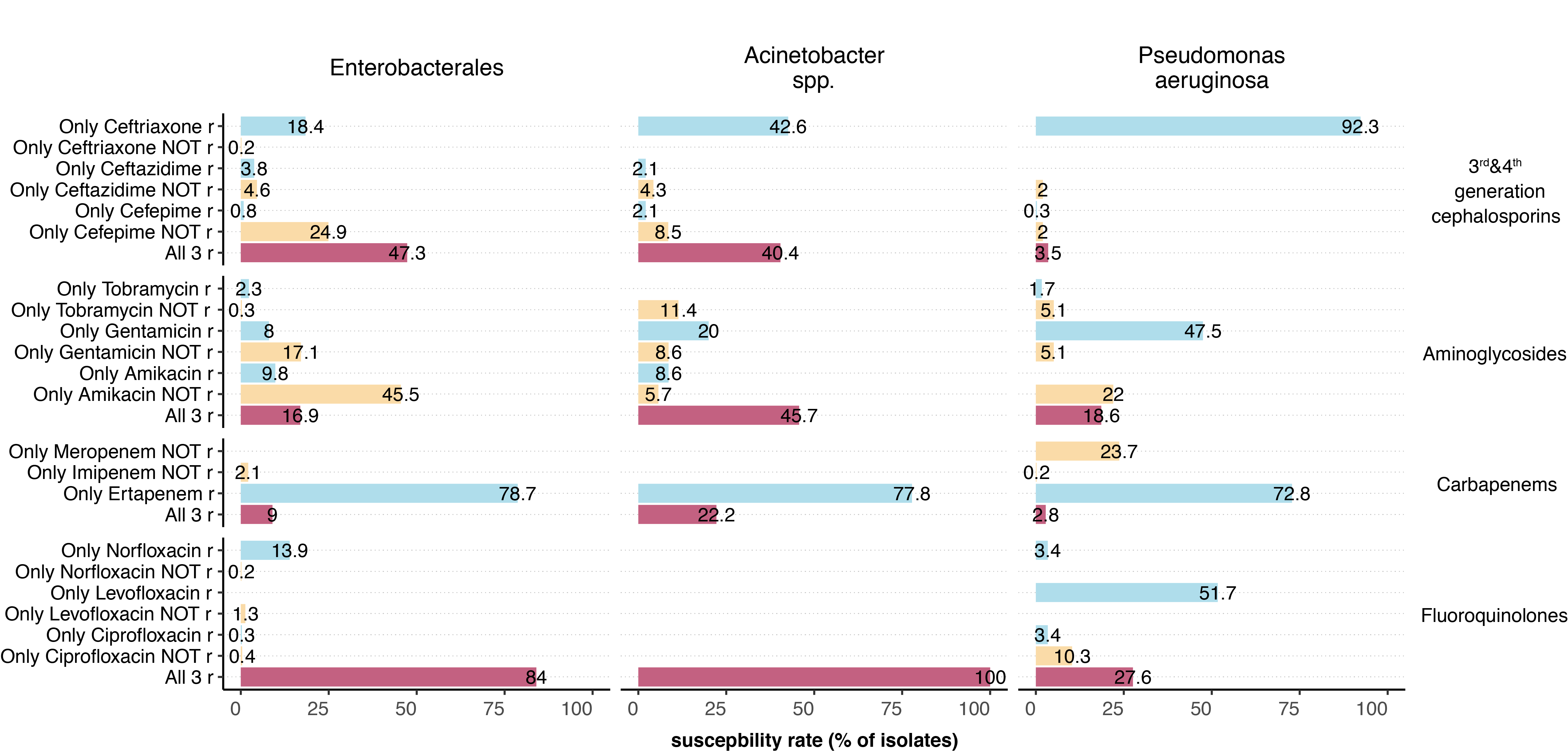
Figure S3 Summary of the cross-resistance in the different gram-negative microorganisms.
To assess cross-resistance within an antibiotic category, only isolates for which all of the most frequently tested antibiotics of a group were analysed were included in the analysis. For the number of isolates included per antibiotic category, see Table S3. Colour code indicates how many antibiotics of the antibiotic category the isolates are resistant (blue, 1 out of 3; orange, 2 out of 3, red, 3 out of 3). r, resistant.
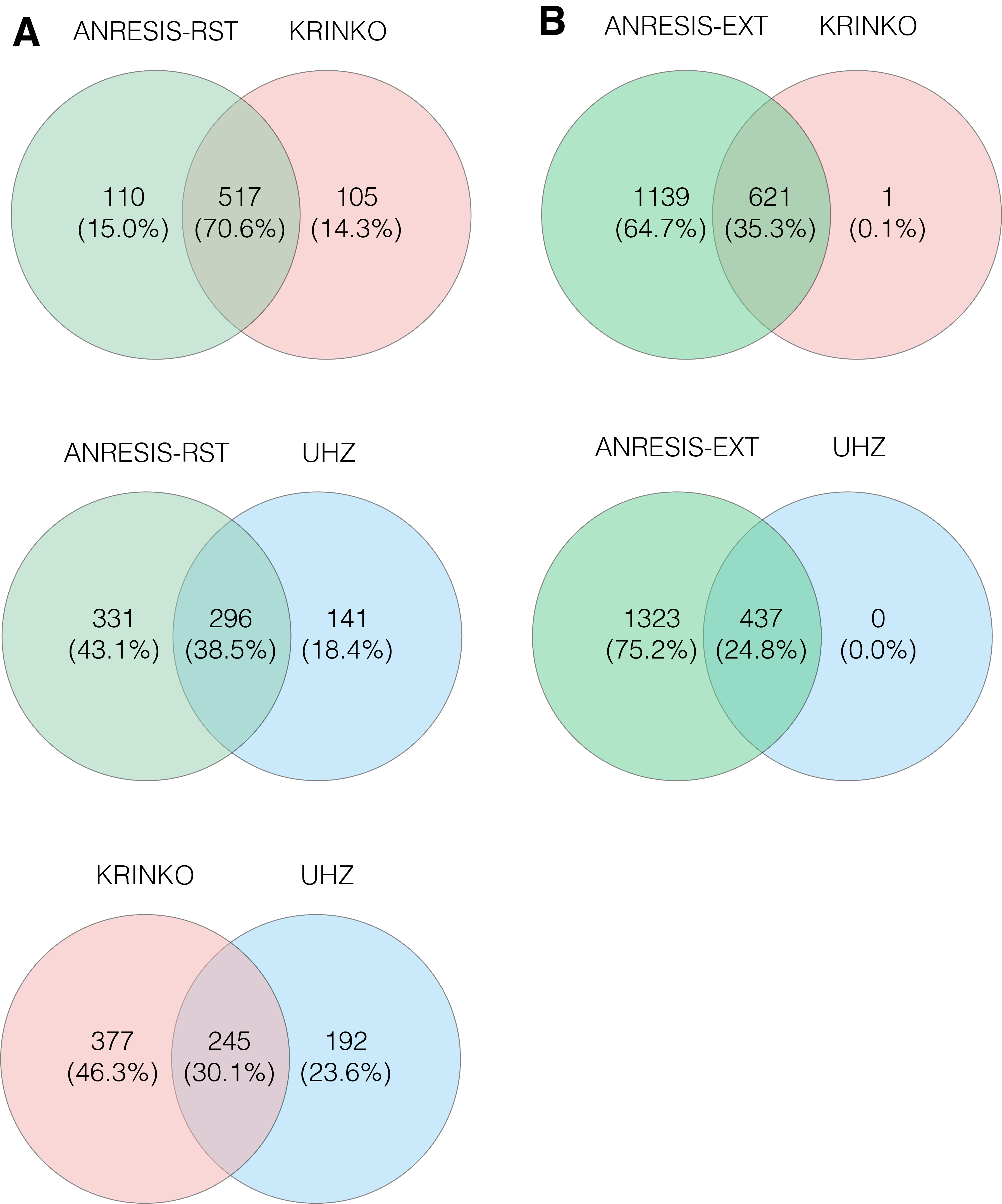
Figure S4 A) Overlap in the number of Enterobacterales isolates classified as MDR-Enterobacterales following the application of the KRINKO, UHZ and ANRESIS-RST definitions.
B) Overlap in the number of Enterobacterales isolates classified as MDR-E following the application of the KRINKO, UHZ and ANRESIS-EXT definitions.
MDR, multiple drug resistance; KRINKO, Commission for Hospital Hygiene and Infection Prevention; UHZ, University Hospital Zurich; ANRESIS, Swiss Centre for Antibiotic Resistance; RST, restricted; EXT, extended
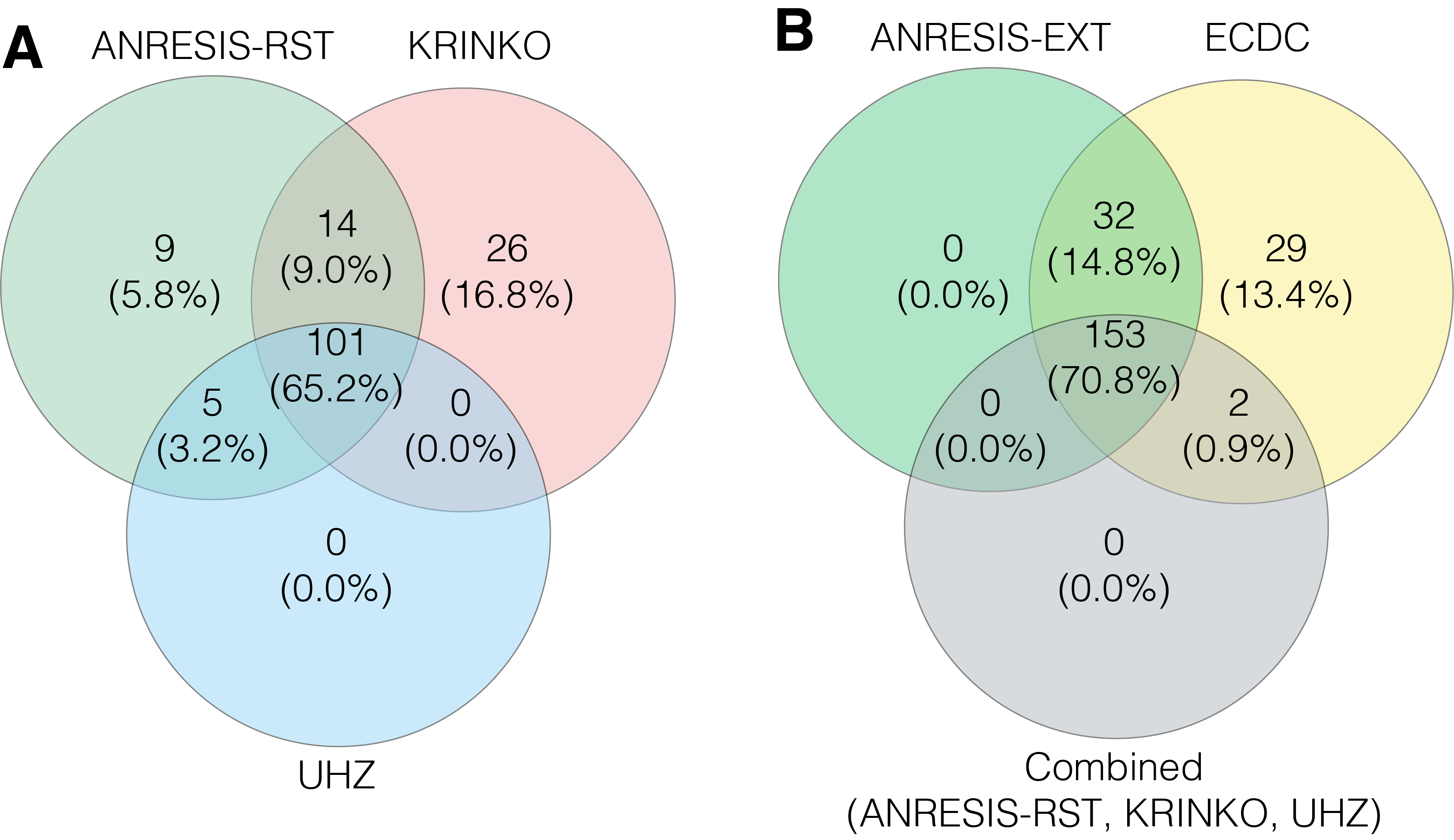
Figure S5 A) Overlap in the number of Pseudomonas aeruginosa isolates classified as MDR following the application of the KRINKO, UHZ and ANRESIS-RST definitions.
B) Overlap in the number of Pseudomonas aeruginosa isolates classified as MDR after application of the ECDC, ANRESIS-EXT definition and the combined MDR cases found with the KRINKO, UHZ and ANRESIS-RST definition.
MDR, multiple drug resistance; ECDC, European Centre for Disease Prevention and Control; KRINKO, Commission for Hospital Hygiene and Infection Prevention; UHZ, University Hospital Zurich; ANRESIS, Swiss Centre for Antibiotic Resistance; RST, restricted; EXT, extended.
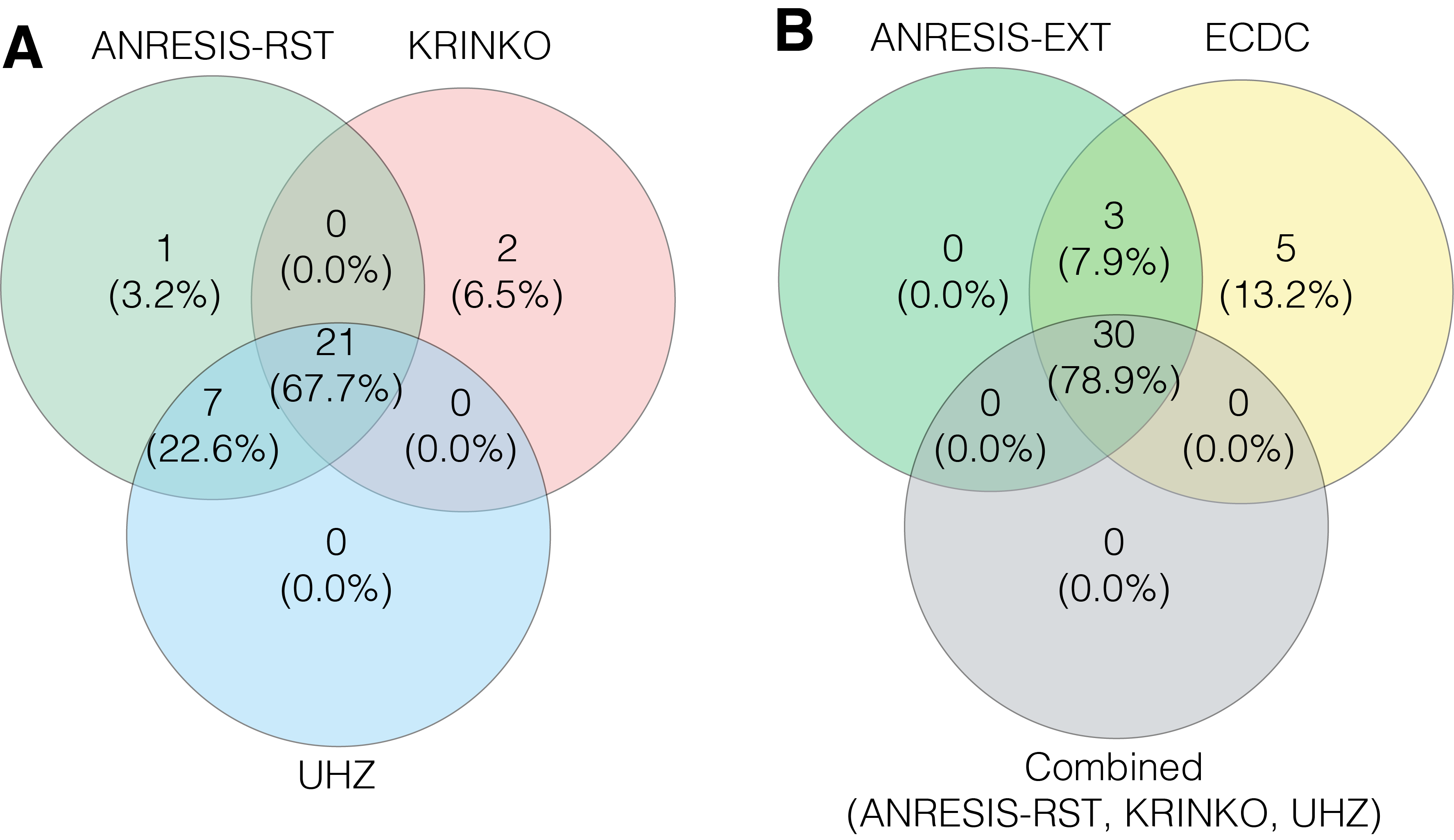
Figure S6 A) Overlap in the number of Acinetobacter spp. isolates classified as MDR following the application of the KRINKO, UHZ and ANRESIS-RST definitions.
B) Overlap in the number of Acinetobacter spp. isolates classified as MDR after application of the ECDC, ANRESIS-EXT definition and the combined MDR cases found with the KRINKO, UHZ and ANRESIS-RST definition.
MDR, multiple drug resistance; ECDC, European Centre for Disease Prevention and Control; KRINKO, Commission for Hospital Hygiene and Infection Prevention; UHZ, University Hospital Zurich; ANRESIS, Swiss Centre for Antibiotic Resistance; RST, restricted; EXT, extended.
...









