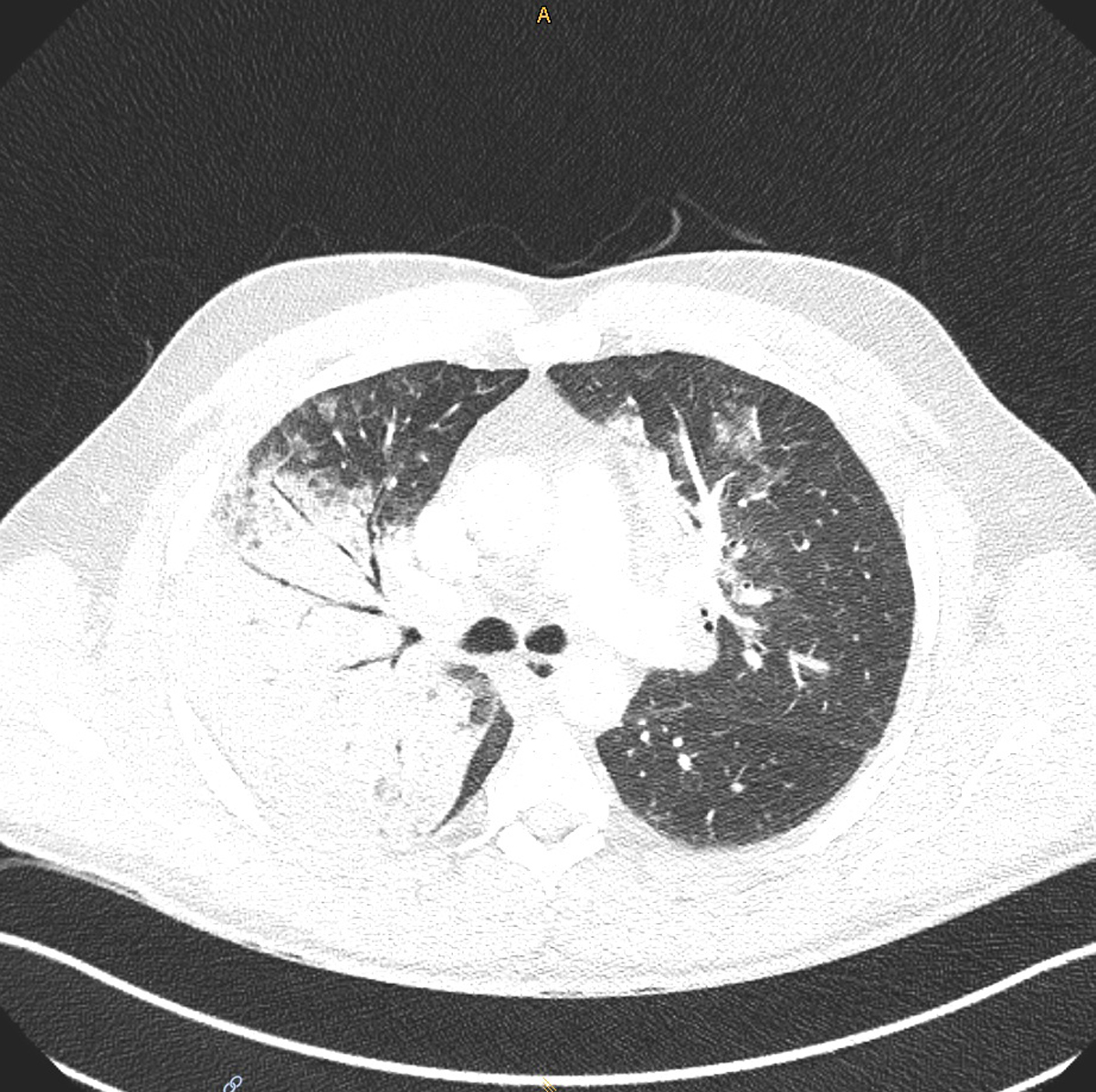
Figure S1 Patient 1: Computed tomography scan of the thorax showing right upper lobe consolidation and bilateral ground glass opacifications (diagnosis: pneumonia due to Chlamydia psittaci).
DOI: https://doi.org/10.4414/SMW.2022.w30102
The unique bacterial phylum Chlamydiae exists all over the world and members of it cause disease not only in humans but also in birds, livestock and wildlife. In humans, Chlamydia trachomatis is the most common bacterial cause of sexually transmitted diseases worldwide and reported numbers of genital C. trachomatis infections are increasing worldwide, including in Switzerland [1, 2]. Ocular strains of C. trachomatis are responsible for the disease trachoma, the most common cause of infectious blindness worldwide and a neglected tropical disease [3]. Chlamydia pneumoniae is a widespread respiratory pathogen in humans and has been detected in many different animal hosts including birds and reptiles [4]. Current research questions address how often C. pneumoniae occurs as co-infection with SARS‑CoV‑2 and if such co-infections can result in more severe disease courses [5].
The Chlamydiaceae family further comprises several zoonotic pathogens, of which the most significant species include Chlamydia psittaci and Chlamydia abortus [6]. C. psittaci is the causative agent of avian chlamydiosis and human psittacosis and is arguably the most important veterinary chlamydial agent in terms of public health and the economy [6]. The current epidemiological situation of human C. psittaci infections is difficult to assess because new comprehensive studies are lacking, and the disease is notifiable in some countries only. In a recent meta-analysis, it was estimated that 1% of the cases of community-acquired pneumonia in the Netherlands were caused by C. psittaci [7]. Worldwide, C. psittaci infections in birds occur in more than 460 free-living or pet bird species [6]. In birds, the infection can be inapparent, severe, acute or chronic with intermittent shedding. To date, 15 genotypes of C. psittaci have been identified, most of which are considered to be readily transmissible to humans. In Switzerland, avian chlamydiosis due to C. psittaci infections is a notifiable disease, with 50 cases in birds reported to the Federal Food Safety and Veterinary Office from 2010 to 2020 [8]. C. psittaci infection in humans can cause fever, respiratory symptoms and life-threatening pneumonia [9]. Therefore, infections with C. psittaci must be ruled out as a differential diagnosis if a patient displaying such symptoms had contact to birds, most notably psittacines and pigeons [6].
C. abortus is a close phylogenetic relative of C. psittaci and causes rampant infectious abortion in sheep and goats, termed ovine enzootic abortion, as well as stillbirths and weak neonates [6]. In Switzerland, C. abortus represents the most common cause of infectious abortion in sheep and goats followed by Coxiella burnetii and Toxoplasma gondii, two other zoonotic pathogens. C. abortus and Coxiella burnetii are both notifiable pathogens in sheep and goats according to the Federal ordinance of epizootic diseases [10]. In humans, C. abortus causes abortion in women and psittacosis-like symptoms in men and women [6].
Despite recent advances, both surveillance and documentation of Chlamydia-related zoonotic cases must be improved to understand the extent and impact of such infections. Knowledge and awareness about the zoonotic potential and the assessment of the exact risk requires collaboration between veterinarians, human health clinicians and microbiologists in a "One Health" setting. Two examples of such an integrated interdisciplinary approach in which professionals from the domains of human health and veterinary health shared methodology and data referring to zoonotic chlamydial infections are presented in this report.
In December 2020, a 52‑year‑old man with an unremarkable medical history presented to his primary care doctor with a 7‑day history of fever and dry cough. Polymerase chain reaction testing (PCR) of a nasopharyngeal swab was negative for SARS‑CoV‑2. Symptoms were treated with acetaminophen and a short-acting bronchodilator. During the following days, self-measured oxygen saturation repeatedly dropped to a minimum of 84%, which prompted the patient to present to the emergency department of a local hospital. Computed tomography (CT) showed bilateral patchy ground-glass opacities as well as right upper lobe consolidation (supplementary fig. S1 in the appendix). Laboratory examination indicated signs of bacterial infection and impaired renal function (table 1). Legionella and pneumococcal antigen in urine and a bacterial broad spectrum PCR from bronchoalveolar lavage fluid were negative. The patient was started on ceftriaxone as empirical therapy for lobar pneumonia and was admitted to the ward. He quickly developed tachypnoea up to 36/min despite supplemental oxygen, a sign of progressive respiratory decompensation, and was therefore transferred to the nearest hospital with available intensive care unit (ICU) capacity the same day. Noninvasive ventilation was commenced, and the antimicrobial regimen was augmented with clarithromycin to cover pathogens causing atypical pneumonia. Since the trial of noninvasive ventilation failed, the patient was eventually intubated endotracheally, and dobutamine and nitroglycerin were administered for a brief period because of a non‑ST elevation myocardial infarction type II and low central venous oxygen saturation. At this time, the intensivists in charge learned from the patient’s wife that he was breeding parrots as a hobby. Thus, due to possible exposure to C. psittaci, antimicrobial therapy with ceftriaxone was supplemented with doxycycline. Further respiratory deterioration (paO2 6.0 kPa on FiO2 1.0, P/F ratio 45 mm Hg) prompted the placement of a veno-venous extracorporeal membrane oxygenation (vv-ECMO) system and the patient was transferred to the University Hospital Zurich, the nearest tertiary care hospital. Antimicrobial therapy was escalated to piperacillin/tazobactam and doxycycline as the patient’s condition rapidly progressed to septic shock, necessitating continuous norepinephrine infusion (30 µg/min). Since the patient developed acute renal failure with acidosis and anuria, continuous veno-venous haemodialysis was initiated. Muscle relaxation (atracurium 60 mg/h) to permit lung protective ventilation (positive end expiratory pressure 15 mbar, driving pressure 14 mbar, plateau pressure 28–30 mbar, tidal volume 4–6 ml/kg) with FiO2 1.0, inhaled nitric oxide (20 ppm) and intermittent prone positioning were applied. In addition, an esmolol infusion (200–300 µg/kg/min) was initiated under careful global haemodynamic monitoring to increase the ratio of ECMO blood flow to cardiac output (QECMO/QCO).
Table 1Patient 1: Blood chemistry results at the local and tertiary care hospital (final diagnosis: pneumonia due to Chlamydia psittaci).
| Laboratory findings | Local hospital a | Tertiary care hospital a | |||
| Patient 1 | Reference range | Patient 1 | Reference range | ||
| C-reactive protein | mg/l | 382 | <10.0 | 272 | <5 |
| Procalcitonin | mg/l | 7.6 | <0.1 | 74.7 | <0.1 |
| Leucocytes | G/l | 5.9 | 3.0–9.6 | 19.15 | 3.0–9.6 |
| Neutrophils | % | 94.9 | 42.2–75.2 | 82.3 | 40–74 |
| Thrombocytes | G/l | 206 | 143–400 | 270 | 143–400 |
| Alkaline phosphatase (AP) | U/l | – | – | 144 | 35–105 |
| Alanine aminotransferase (GPT) | U/l | 50 | 10–50 | 72 | <35 |
| Aspartate aminotransferase (GOT) | U/l | 105 | 10–50 | 277 | <35 |
| Gamma-glutamyltransferase (GGT) | U/l | – | 5–36 | 147 | <40 |
| Lactate dehydrogenase (LDH) | U/l | 506 | 135–225 | 1293 | 240–480 |
| Bilirubin | mmol/l | 10.1 | <21 | 12 | <21 |
| D-dimer | mg/l | – | – | 5.68 | < 0.5 |
| Sodium | mmol/l | 130 | 136–145 | 140 | 136–145 |
| Potassium | mmol/l | 3.9 | 3.6–5.5 | 5.1 | 3.4–4.5 |
a Values refer to the day of patient admission at the indicated hospital
At the same time, PCR on a sample taken by the veterinarian from one of the patient’s parrots with nonspecific clinical signs was positive for C. psittaci. In the hospitalised patient, microbiological sampling was repeated, including multiple blood cultures and bronchoscopy with lavage. A multiplex PCR panel specific for pathogens causing atypical pneumonia performed on a bronchial lavage fluid sample from the patient was also positive for C. psittaci. Serology revealed IgA and IgG titres consistent with an active C. psittaci infection. An overview of C. psittaci-related molecular and serological findings is shown in table 2, the corresponding methods details are outlined in the appendix. Additional microbiological and serological analyses are summarised in table S1 (appendix).
Table 2Patient 1: PCR and serology results related to Chlamydia psittaci infection.
| Day a | Sample type | PCR result b | Mean cycle threshold value | Serology (test/titre) | |
| 2 | Bronchoalveolar lavagec | C. psittaci positive | 30.7 | ||
| 3 | Serum | C. psittaci positived | 27.23 | C. psittaci (IgG) | 1:128 |
| C. psittaci (IgA) | 1:64 | ||||
| Tracheobronchial secretion | C. psittaci positive | 20.91 | |||
| 4 | Serum | C. psittaci positived | 26.48 | C. psittaci (IgG) | 1:512 |
| C. psittaci (IgA) | 1:64 | ||||
PCR: polymerase chain reaction
a Refers to time point of sample collection after admission to tertiary care hospital (University Hospital Zurich) unless otherwise stated
b Multiplex PCR panel for respiratory pathogens causing atypical pneumonia (see materials and methods in the appendix)
c Sample was already taken at the local hospital for bacterial broad spectrum PCR analysis, but was re-analysed with the respiratory Multiplex PCR panel two days after patient’s admission to the University Hospital Zurich.
d Retrospective analysis outside of routine diagnostic procedure
Eleven days after initial onset of symptoms, bilateral, widened pupils non-reactive to light were noted on routine physical examination. CT scan of the brain revealed a large intra-parenchymal haemorrhage located in the right frontal lobe with consequent compression of the ventricles, transtentorial and subfalcine herniation, and generalised cerebral oedema (supplementary fig. S2). Predisposing factors for such catastrophic bleeding were thrombocytopenia (Nadir 37 G/l), owed to extracorporeal therapies (vv-ECMO and CVVHD), necessary anticoagulation and sepsis. Notably, activated clotting time was in the recommended range of 150–180 sec. In consideration of septic multi-organ failure and a futile neurological prognosis, the treating clinicians together with the patient’s family decided to shift from medical to palliative care and the patient passed away after withdrawal of ECMO.
As mentioned above, after the attending physician contacted the veterinarian and asked whether zoonotic agents contracted from birds should be considered as a differential diagnosis, a Senegal parrot (Poicephalus senegalus) with a nonspecific clinical presentation such as weight loss and an enlarged liver from the holding of patient 1 was sampled at the veterinary clinic. This eyelid-choana-cloaca-swab simultaneously tested positive for Chlamydiaceae and C. psittaci by PCR (supplementary table S2 in the appendix). This result was communicated to the physician and the cantonal veterinary authority, as chlamydiosis is a notifiable animal disease [10]. The holding of patient 1 contained over 100 psittacines of various species, 13 chickens and 15 quails; kept in cages or aviaries (table S2). The veterinary authorities banned all animal traffic in the holding and conducted a comprehensive follow-up examination. Genotyping of selected samples (including bronchial lavage fluid of patient 1) revealed C. psittaci genotype A (table S2, detailed method description in the appendix). Larger parrots kept in cages were weekly treated with doxycycline i.m. over the course of 45 days, all aviary birds received doxycycline via their drinking water for 45 days. The premises were cleaned and disinfected twice (potassium peroxymonosulfate; Virkon™ S, Lanxess, Sudbury, UK) by a professional company. A first disinfection check was performed in April, with 2/19 samples positive for C. psittaci by PCR (table S2). After a second round of disinfection in May, 2/7 environmental samples were still positive in aviary 1, prompting a second round of treatment of the birds. After a final disinfection check in September with 7/7 samples testing negative, the ban on the premises was lifted by the authorities.
In April 2020, a 34-year-old pregnant (week 25) female was admitted to a local hospital because of a persistent dry cough, fever of up to 40 ºC, occasional chills, fatigue and headache for 4 days. Apart from a mild form of hyperemesis gravidarum, the pregnancy was so far without complications. Laboratory examination upon admission revealed increased levels of inflammatory markers (table 3, left panel). In addition, levels of thrombocytes, lactate dehydrogenase and liver enzymes suggested a probable HELLP syndrome (Haemolysis, Elevated Liver enzymes, and Low Platelet count). The patient displayed slight hyponatraemia and hypokalaemia. Initial screening for SARS‑CoV‑2, influenza A/B and respiratory syncytial virus were negative (supplementary table S3 in the appendix). Lung ultrasound showed ubiquitous, non-confluent b‑lines and no signs of pleural effusion. Abdominal ultrasonography revealed neither abnormalities of the liver nor cholestasis. The patient was administered empirically intravenous amoxicillin / clavulanic acid and anticoagulation was started. In addition, potassium substitution therapy was initiated. Twenty‑four hours after admission, the patient’s thrombocytopenia had become more pronounced and inflammatory makers remained high (table 3, middle panel). Parameters indicative of HELLP syndrome were still significantly elevated, thus the patient was transferred to the ICU of the University Hospital Zurich, a tertiary care hospital. Upon admission, fetal lung maturation with corticosteroids was initiated and magnesium sulphate was administered for fetal neuroprotection. Amoxicillin / clavulanic acid therapy was continued and augmented by intravenous clarithromycin. A CT scan of the thorax revealed pulmonary infiltrations in the left upper lobe as well as bilateral lower lobe infiltrates, but no indication of lung embolism. HELLP syndrome was finally excluded, as levels of liver enzymes decreased continuously (table 3, right panel).
Table 3Patient 2: Blood chemistry results at the local and tertiary hospital (final diagnosis: pneumonia due to Chlamydia abortus).
| Laboratory findings | Local hospital | Tertiary care hospital a | ||||
| Patient 2 (day 1) | Patient 2 (day 2) | Reference range | Patient 2 | Reference range | ||
| C-reactive protein | mg/l | 109.3 | 133.8 | <5.0 | – | – |
| Procalcitonin | mg/l | 3.2 | 5.2 | <0.1 | – | – |
| Leucocytes | G/l | 6.4 | 5.7 | 3.9–10 | – | – |
| Neutrophils | % | 81.9 | 80.6 | 34.0–71.0 | – | – |
| Thrombocytes | G/l | 101 | 70 | 182–369 | 294 | 143–400 |
| Alkaline Phosphatase (AP) | U/l | – | 261 | 35–104 | 179 | 35–105 |
| Alanine aminotransferase (GPT) | U/l | 197 | 167 | <33 | 47 | <35 |
| Aspartate aminotransferase (GOT) | U/l | – | 143 | <33 | 22 | <35 |
| Gamma glutamyltransferase (GGT) | U/l | 105 | 99 | 5–36 | 107 | <40 |
| Lactate dehydrogenase (LDH) | U/l | 648 | 642 | 240–480 | 380 | 240–480 |
| Bilirubin | mmol/l | 22.9 | 35.6 | <17 | 11 | <21 |
| D-dimer | mg/l | 4.4 | – | <0.5 | – | – |
| Sodium | mmol/l | 131 | 131 | 136–149 | – | – |
| Potassium | mmol/l | 2.95 | 3.25 | 3.6–5.5 | – | – |
a Values on day of the patient discharge from tertiary care hospital (University Hospital Zurich)
The patient was therefore transferred to the normal prenatal ward. Routine bacteriological tests including urine and blood cultures were negative. Finally, C. abortus was detected a in throat swab using a two-step PCR / melting curve analysis approach (table 4; for a detailed description of the PCR test, see the appendix). Serology revealed high IgG and IgA titres for C. pneumoniae (table 4). However, whether this finding could be attributed to cross-reactivity caused by the C. abortus infection or a previous C. pneumoniae infection remained unclear. Additional microbiological and serological analyses are summarised in table S3. Based on the identification of C. abortus, therapy with amoxicillin / clavulanic acid was stopped and clarithromycin was changed to oral administration. Finally, PCR analysis of sputum was negative for C. abortus 6 days after clarithromycin had been started. The patient was discharged home in a stable general condition 9 days after admission. Oral clarithromycin (500 mg twice a day) was continued for another 4 days.
Table 4Patient 2: PCR and serology findings related to Chlamydia abortus infection.
| Day a | Sample type | Respiratory PCR result b | Mean cycle threshold value | Serology (test/titre) | |
| 1 | Blood culture (aerobic) c | C. abortus positive | 36.54 | ||
| Blood culture (anaerobic)c | C. abortus negative | ||||
| Serumc | C. psittaci (IgG) | 1:1281 | |||
| C. psittaci (IgA) | 1:161 | ||||
| C. pneumoniae (IgG) | 1:20,481 | ||||
| C. pneumoniae (IgA) | 1:1024 | ||||
| 2 | Throat swab | C. abortus positive | 34.52 | ||
| 5 | Throat swab | Negative | |||
| 7 | Blood culture (aerobic)c | Negative | |||
| Blood culture (anaerobic) c | Negative | ||||
| 9 | Sputum | Negative | |||
PCR: polymerase chain reaction
a Refers to time point of sample collection after admission to tertiary care hospital (University Hospital Zurich)
b Multiplex PCR panel for respiratory pathogens causing atypical pneumonia (see appendix)
c Retrospective analysis outside of routine diagnostic procedure
A more detailed retrospective inquiry of the patient revealed that she was living on a farm with a flock of about 30 sheep and 6 goats. After an episode of massively increased numbers of abortions among ewes about 6 years ago, all sheep were vaccinated against C. abortus. However, in March 2019 two unvaccinated goats both aborted twins. These were subsequently handled and removed by the patient, making a pulmonary infection via C. abortus-harbouring aerosols a likely scenario. Finally, it remained unclear whether veterinary authorities were involved in the case for follow-up investigations as the patient preferred not to comment on that.
Since 2019/2020, the SARS‑CoV‑2 overwhelms our population with human pneumonia cases associated with this emerging virus. Despite this, other pathogenic agents including zoonotic bacteria should be considered as differential diagnosis for pneumonia, in particular when a COVID test remains negative and previous animal contact is reported by the patient. This animal contact might be traced back to livestock husbandry (e.g., sheep and goat farming with breeding activities) or pet housing (e.g., bird handling and trading). It is important to ask the patient or their relatives for potential risk factors, including questions about contact with mammals, birds and reptiles during the clinical history evaluation. Interdisciplinary communication between the patient’s physician and the veterinarian in charge of the respective animal husbandry/keeping or even the cantonal veterinary authorities is highly recommended and might establish the missing link. Diagnostic methods for animal pathogens might not be readily available in medical microbiology laboratories and samples in question might be referred to specialised veterinary diagnostic laboratories, some of which also serve as national reference laboratories for notifiable diseases in animals (https://www.blv.admin.ch/blv/de/home/tiere/tierseuchen/tierseuchendiagnostik.html). Whereas chlamydial abortion in sheep and goats due to C. abortus and avian Chlamydiosis in birds due to C. psittaci are notifiable diseases in Switzerland and worldwide, the zoonotic counterparts are not, at least in Switzerland. Increased surveillance and awareness of such zoonotic cases is warranted and the two case reports presented in this study aim to do so.
All authors have completed and submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest. No potential conflict of interest was disclosed.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
The laboratory work was partly performed using the logistics of the Centre for Clinical Studies at the Vetsuisse Faculty of the University of Zurich. We would like to thank Theresa Pesch, Barbara Prähauser, Brigitte Sigrist and Petra Bruggmann for technical support, as well as the co-workers of the Molecular Diagnostics Department and the Serology Department at the Institute of Medical Microbiology.

Figure S1 Patient 1: Computed tomography scan of the thorax showing right upper lobe consolidation and bilateral ground glass opacifications (diagnosis: pneumonia due to Chlamydia psittaci).
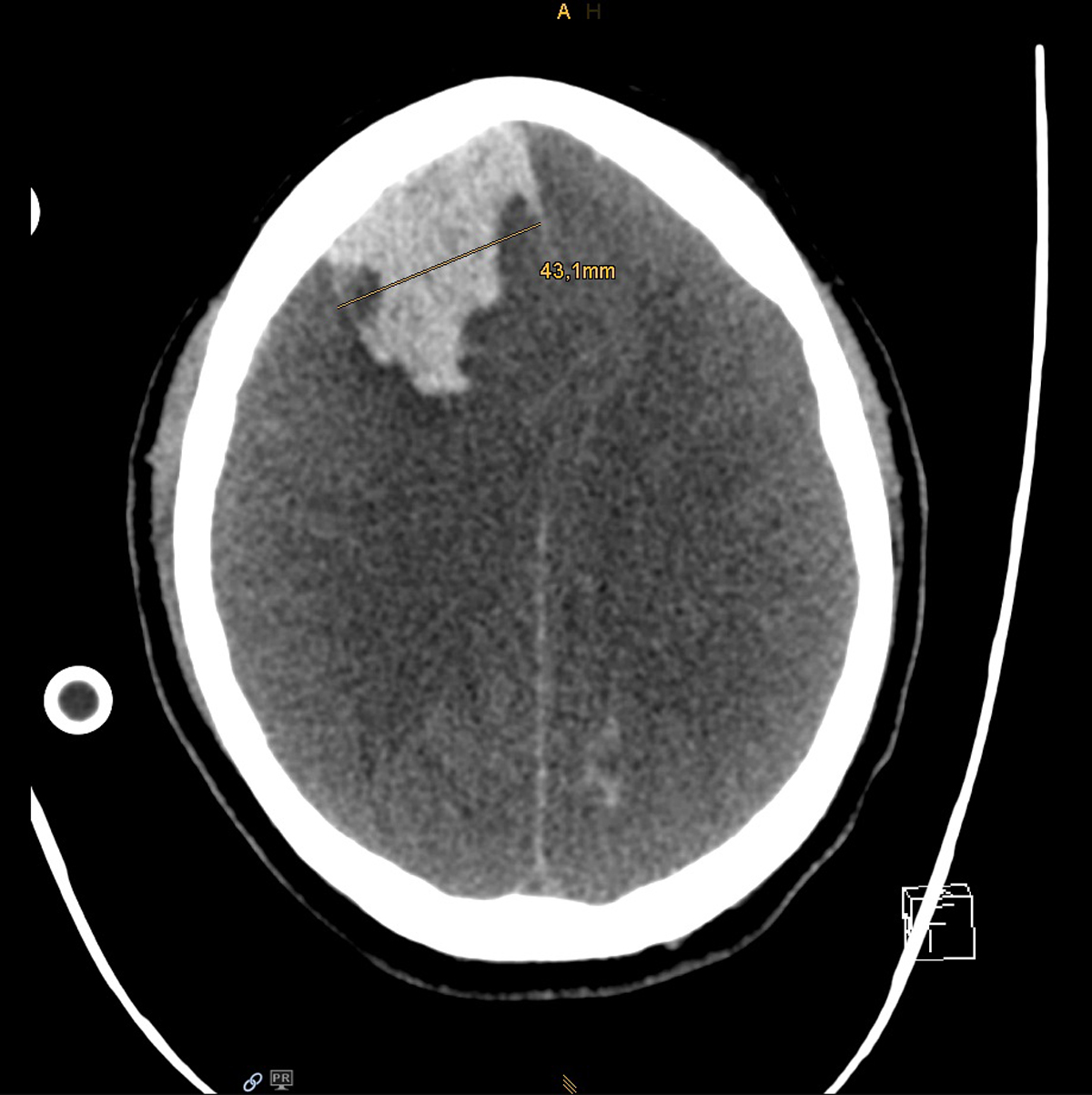
Figure S2 Patient 1: Computed tomography scan of the brain showing acute intraparenchymal bleeding in the right frontal lobe and generalised cerebral oedema.
Table S1Patient 1: Additional diagnostic findings (final diagnosis: pneumonia due to Chlamydia psittaci).
| Method | Sample type | Performed tests | Result |
| PCR | Nasopharyngeal swab | SARS-CoV-2 | Negative |
| Bronchoalveolar lavage | Bacterial broadspectrum PCR | Negative | |
| Culture | Blood | No growth | |
| Bronchoalveolar lavage | No growth | ||
| Tracheobronchial secretion | No growth | ||
| Serology | Bronchoalveolar lavage | Aspergillus antigen | Negative |
| Other | Urine | Legionella antigen | Negative |
| Streptococcus pneumoniae antigen | Negative |
Table S2Detailed results of different molecular tests using a step-wise approach for the detection and identification of Chlamydia in human (patient 1 and 2) and bird samples from Switzerland.
| Case No. | Sample | Chlamydiaceae qPCR (23S rRNA) | C. psittaci qPCR (ompA) 1 | ompA genotyping PCR | Microarray assay (23S rRNA) | 16S rRNA PCR and sequencing (% sequence identity) | |||
| Sample origin | Day of sampling | Sample type | Ct values (duplicates) | Number of copies per µl DNA | |||||
| 1 | Parrot cage: Senegal parrot 1 2 | 22 Dec 2020 | Eyelid-CSS | 29.6 / 29.4 | 3.02E+02 | 1 / 1 pos | genotype A | n.d. | n.d. |
| 1 | Parrot cage: Senegal parrot 2 | 24 Dec 2020 | CSS | 24.7 / 24.9 | 7.53E+03 | 1 / 1 pos | genotype A | n.d. | n.d. |
| 1 | Parrot cage: Senegal parrots 3 and 4 | 24 Dec 2020 | CSS | n.d. | n.a. | 2 / 2 pos | n.d. | n.d. | n.d. |
| 1 | Aviary 1: quails | 24 Dec 2020 | Pooled CSS | 31.7 / 31.8 | 6.32E+01 | 1 / 1 pos | genotype A | n.d. | n.d. |
| 1 | Aviary 1: psittacines | 24 Dec 2020 | Pooled CSS | 27.2 / 27.0 | 1.55E+03 | 1 / 1 pos | genotype A | n.d. | n.d. |
| 1 | Aviary 1: psittacinesand chickens | 24 Dec 2020 | Pooled CSS | n.d. | n.a. | 3 / 3 pos | n.d. | n.d. | n.d. |
| 1 | Aviary 1: environment | 24 Dec 2020 | Dust (dander) | n.d. | n.a. | 2 / 2 pos | n.d. | n.d. | n.d. |
| 1 | Aviary 2: environment | 24 Dec 2020 | Dust (dander) | n.d. | n.a. | 5 / 5 neg | n.d. | n.d. | n.d. |
| 1 | Aviary 1: disinfection control April | 08 Apr 2021 | Various swabs | n.d. | n.a. | 1 / 10 pos | n.d. | n.d. | n.d. |
| 1 | Aviary 2: disinfection control April | 08 Apr 2021 | Various swabs | n.d. | n.a. | 7 / 7 neg | n.d. | n.d. | n.d. |
| 1 | aparrot cage: Senegal parrots2 and 4 3 | 082021 | CSS | n.d. | n.a. | 1 / 2 pos | n.d. | n.d. | n.d. |
| 1 | Aviary 1: disinfection control May | 04May 2021 | Various swabs | n.d. | n.a. | 2 / 7 pos | n.d. | n.d. | n.d. |
| 1 | Parrot cage: Senegal parrots2 and 4 | 04 June 2021 | CSS | n.d. | n.a. | 2 / 2 neg | n.d. | n.d. | n.d. |
| 1 | Aviary 1: disinfection control Sept | 08 Sept 2021 | Pooled faeces | n.d. | n.a. | 7 / 7 neg | n.d. | n.d. | n.d. |
| 1 | Patient 1 | 22 Dec 2020 | BAL | 22.1 / 22.0 | 1.29E+04 | n.d. | genotype A | n.d. | n.d. |
| 1 | Patient 1 | 242020 | Serum | 30.7 / 30.8 | 3.13E+01 | n.d. | no result | n.d. | n.d. |
| 1 | Patient 1 | 252020 | Serum | 38.9 / 39.3 | 1.02E-01 | n.d. | no result | n.d. | n.d. |
| 2 | Patient 2 | 13 Apr 2020 | Throat swab | 39.71 / 43.97 | 0.1 | n.d. | n.a. | C. abortus | C. abortus (100%) |
| 2 | Patient 2 | 122020 | Blood | Undet / undet | n.a. | n.d. | n.a. | n.d. | n.d. |
BAL: bronchoalveolar lavage; CCS: choanal-cloacal swab; Ct: cycle threshold; n.d.: not done; n.a.: not applicable; PCR: polymerase chain reaction; pos: positive; neg: negative; undet: undermined (negative)
1 Number of samples positive per tested individual or pooled samples.
2 One out of four birds (No. 1) died one day after sampling.
3 Whilst treating, one of the birds (No. 3) died because of a fight with other parrots.
Table S3Patient 2: Additional diagnostic findings (final diagnosis: pneumonia due to Chlamydia abortus).
| Method | Material | Performed tests | Result | Reference range |
| PCR | Nasopharyngeal swab | SARS-CoV-2 | Negative | |
| Influenza A and B | Negative | |||
| Respiratory syncytial virus | Negative | |||
| Urine | C. trachomatis and Neisseria gonorrhoeae | Negative | ||
| Culture | Sputum | No growth | ||
| Cervical swab | Normal flora | |||
| Urine, first portion | No growth | |||
| Blood | No growth | |||
| Serology | Serum | HIV AK/AG Combination | Negative | |
| CMV IgG | 21 | ≥6 U/ml | ||
| CMV IgM | Negative | |||
| EBV VCA IgG | Positive | |||
| EBV VCA IgM | Negative | |||
| HSV-1/2 IgG | >1000 | >30 U/ml | ||
| HSV-1/2 IgM | Borderline | |||
| Parvovirus B19 IgG | Positive | |||
| Parvovirus B19 IgM | Negative | |||
| Rubella IgG | 50.2 | ≥10 U/ml | ||
| Rubella IgM | Negative | |||
| Toxoplasma IgG | 0.2 | ≥3 U/ml | ||
| Toxoplasma IgM | Negative | |||
| Treponema pallidum TPPA | Negative | |||
| SARS-CoV-2 IgG | Negative | |||
| ARS-CoV-2 IgM | Negative |
Automated DNA extraction from patient samples was performed using the QIAsymphony platform together with the QIASymphony DSP Virus/Pathogen Kit. From veterinary and environmental samples as well as patient samples that were subjected to C. psittaci ompA genotyping (see below), DNA was extracted as described previously [11] using the Maxwell 16 Buccal Swab LEV DNA Purification Kit (Promega, Madison, WI, USA) adhering to instructions provided by the manufacturer. DNA concentration and quality were determined using Nanodrop-1000 (Witec AG, Lucerne, Switzerland).
Detection of bacterial pathogens causing atypical pneumonia from respiratory and non-respiratory samples was performed using a Lighmix®-based (TIBMolBiol, Berlin, Germany) multiplex PCR panel comprising C. pneumoniae / C. psittaci, Bordetella pertussis/parapertussis, Mycoplasma pneumoniae and Legionella pneumophila [12]. Samples that were positive for C. pneumoniae / C. psittaci were re-tested by singleplex PCRs in order to distinguish the two species. Finally, C. psittaci-positive samples were subjected to a melting curve analysis, which differentiates between C. psittaci, C. abortus and C. caviae.
Initial screening with the Chlamydiaceae family-specific qPCR was performed as described previously [11]. Specifically, this qPCR targets a 111 bp segment of the 23S ribosomal RNA (rRNA) [13], and was performed on an ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA). An internal amplification control (117 bp) was included using Intype IC-DNA as described [11]. All samples were tested in duplicate with a cycle threshold value of 0.1 set after each run. A cycle threshold (Ct value) <38 was considered positive. A C. abortus standard curve ranging from 10 to 107 copies was used for quantification and a water sample served as a negative control.
The average number of copies per sample and qPCR was determined using the standard formula (1): Y = Ae bx (1) [A = y‑intercept, e = mathematical constant (2.71828), b = exponential growth constant Ct values (y) and corresponding copy number (x) of the standard curves were used to calculate A and b for each run]. Two Ct values per sample, qPCR and run were then used to calculate the average number of copies per µl of sample.
The species-specific PCR was performed according to the protocol described previously [14]. The reaction mix contained 4 μl (<150 ng/µl) sample template, 1 µl eGFP template, 1x TaqMan Universal PCR MasterMix, 900 nM of the primers CppsOMP1‑F and CppsOMP1‑R, 200 nM probe CppsOMP1‑S, 900 nM of the primers eGFP‑1‑F and eGFP‑2‑R and 200 nM probe eGFP‑HEX [15] in a final volume of 25 μl.
Chlamydiaceae-positive qPCR samples were subjected to a short 16S rRNA pan Chlamydiales PCR followed by Sanger sequencing. DNA amplification of the 298 bp product was performed with the Biometra TRIO thermal cycler (Analytik Jena AG) according to the slightly modified protocol of [16]. Hence, each reaction consisted of 3 μl DNA template, 25 μl AmpliTaq GoldTM 360 Master Mix (Thermo Fisher Scientific), 19 μl molecular grade water and 1.5 μl of 16S IGF (forward) and 16S IGR (reverse) primers (10 μM, Microsynth) adding up to a total volume of 50 μl. Cycling conditions consisted of initialisation at 95 °C for 10 min followed by 40 cycles of 95 °C for 30 s, 58 °C for 30 s and 72 °C for 60 s as well as a final extension at 72 °C for 7 min. Positive and negative controls were included in each run. PCR products were separated by agarose gel electrophoresis and PCR-positive samples were prepared for Sanger sequencing.
Next, all positive samples were investigated with the Arraymate microarray, which targets a multivariable sequence on the 23S rRNA gene (Alere, Jena, Germany) allowing identification of 12 twelve Chlamydiaceae species: C. abortus, C. avium, C. caviae, C. felis, C. gallinaceae, C. ibidis, C. muridarum, C. pecorum, C. pneumoniae, C. psittaci, C. suis and C. trachomatis, as well as the identification of mixed infections [17]. This array is based on hybridisation of DNA first amplified and biotin-labelled by PCR with the following thermocycler protocol: 96 °C 10 min, 40 cycles of 94 °C 30 s, 50 °C 30 s and 72 °C 30 s. An internal control DNA was included, as recommended by the manufacturer (Intype IC-DNA, Qiagen Labor, Leipzig) [11].
C. psittaci-positive samples were further genotyped by sequencing of the ompA gene. Per sample, a reaction mix with a final volume of 50 μl containing 25 μl REDTaq ReadyMix (Merck KGaA, Darmstadt, Germany), 200 nM of the primers ompA F (CTU) and ompA rev [18], and 3 μl sample template with a DNA concentration of 25 ng/μl was prepared. Cycling conditions were 10 min at 95 °C, followed by 35 cycles of 95 °C for 30 s, 49 °C for 30 s, 72 °C for 60 s, and a final elongation at 72 °C for 7 min [18]. Analysis of ompA nucleotide sequences was conducted using Geneious version 10.2 (Biomatters Ltd., available from https://www.geneious.com).
IgG and IgA antibody titres for Chlamydia sp. were routinely determined using an microimmunofluorescence assay according to the manufacturer’s instructions (Focus Diagnostics).
1. Lanjouw E , Ouburg S , de Vries HJ , Stary A , Radcliffe K , Unemo M . Background review for the ‘2015 European guideline on the management of Chlamydia trachomatis infections’. Int J STD AIDS. 2015 Nov; :0956462415618838. https://doi.org/10.1177/0956462415618838
2. Chlamydiose in der Schweiz im Jahr 2018. In: HIV, Syphilis, Gonorrhoe und Chlamydiose in der Schweiz im Jahr 2018: eine epidemiologische Übersicht. Bundesamt für Gesundheit (BAG). Bulletin. 2019 Oct;41 . [cited 2021 Jun 11] Available from: https://www.bag.admin.ch/bag/de/home/krankheiten/krankheiten-imueberblick/chlamydiose.html
3. Taylor HR , Burton MJ , Haddad D , West S , Wright H . Trachoma. Lancet. 2014 Dec;384(9960):2142–52. https://doi.org/10.1016/S0140-6736(13)62182 https://doi.org/10.1016/S0140-6736(13)62182-0
4. Roulis E , Polkinghorne A , Timms P . Chlamydia pneumoniae: modern insights into an ancient pathogen. Trends Microbiol. 2013 Mar;21(3):120–8. https://doi.org/10.1016/j.tim.2012.10.009
5. De Francesco MA , Poiesi C , Gargiulo F , Bonfanti C , Pollara P , Fiorentini S , et al. Co-infection of chlamydia pneumoniae and mycoplasma pneumoniae with SARS-CoV-2 is associated with more severe features. J Infect. 2021 Apr;82(4):e4–7. https://doi.org/10.1016/j.jinf.2021.01.009
6. Sachse K , Borel N . Recent Advances in Epidemiology, Pathology and Immunology of Veterinary Chlamydiae, in: Tan, M., Hegeman, J.H., Sütterlin, C. (Eds.), Chlamydia Biology: From Genome to Disease. Caister Academic Press, Norfolk, pp. 403–428. 2020; doi:https://doi.org/10.21775/9781912530281.17
7. Hogerwerf L , DE Gier B , Baan B , VAN DER Hoek W . Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2017 Nov;145(15):3096–105. https://doi.org/10.1017/S0950268817002060
8. BLV . (Bundesamt für Lebensmittelsicherheit und Veterinärwesen, Federal Food Safety and Veterinary Office), 2021. Informationssystem Seuchenmeldungen InfoSM, 2021. Available: https://www.infosm.blv.admin.ch [Accessed June 11, 2021].
9. Knittler MR , Sachse K . Chlamydia psittaci: update on an underestimated zoonotic agent. Pathog Dis. 2015 Feb;73(1):1–15. https://doi.org/10.1093/femspd/ftu007
10. Federal ordinance of epizootic diseases (Tierseuchenverordnung), SR 916.401, Available: https://www.fedlex.admin.ch/eli/cc/1995/3716_3716_3716/de [Accessed June 11, 2021].
11. Hoffmann K , Schott F , Donati M , Di Francesco A , Hässig M , Wanninger S , et al. Prevalence of Chlamydial Infections in Fattening Pigs and Their Influencing Factors. PLoS One. 2015 Nov;10(11):e0143576. https://doi.org/10.1371/journal.pone.0143576
12. Wagner K , Springer B , Imkamp F , Opota O , Greub G , Keller PM . Detection of respiratory bacterial pathogens causing atypical pneumonia by multiplex Lightmix® RT-PCR. Int J Med Microbiol. 2018 Apr;308(3):317–23. https://doi.org/10.1016/j.ijmm.2018.01.010
13. Ehricht R , Slickers P , Goellner S , Hotzel H , Sachse K . Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol Cell Probes. 2006 Feb;20(1):60–3. https://doi.org/10.1016/j.mcp.2005.09.003
14. Pantchev A , Sting R , Bauerfeind R , Tyczka J , Sachse K . New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet J. 2009 Aug;181(2):145–50. https://doi.org/10.1016/j.tvjl.2008.02.025
15. Hoffmann B , Depner K , Schirrmeier H , Beer M . A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J Virol Methods. 2006 Sep;136(1-2):200–9. https://doi.org/10.1016/j.jviromet.2006.05.020
16. Everett KD , Bush RM , Andersen AA . Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999 Apr;49(Pt 2):415–40. https://doi.org/10.1099/00207713-49-2-415
17. Schnee C , Sachse K . DNA microarray-based detection of multiple pathogens: mycoplasma spp. and Chlamydia spp. Methods Mol Biol. 2015;1247:193–208. https://doi.org/10.1007/978-1-4939-2004-4_15
18. Sachse K , Laroucau K , Hotzel H , Schubert E , Ehricht R , Slickers P . Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiol. 2008 Apr;8(1):63. https://doi.org/10.1186/1471-2180-8-63