HCV RNA quantification in capillary dried blood spots with the Xpert® HCV Viral Load test for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection
DOI: https://doi.org/10.4414/SMW.2021.w30089
Andrea
Bregenzera, Cornelia
Ottigerb, Cornelia
Krismera, Karin
Sagera, Christoph A.
Fuxa
aDepartment of Infectious Diseases and Hospital Hygiene, Cantonal Hospital Aarau, Switzerland
bInstitute for Laboratory Medicine, Cantonal Hospital Aarau, Switzerland
Summary
BACKGROUND: For patients with difficult venous access after long-term intravenous drug use, rapid point-of-care hepatitis C virus (HCV) RNA quantification in capillary whole blood with the Xpert® HCV Viral Load Fingerstick (VL FS) test (60 minutes) is a convenient and reliable method for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection. However, an expensive GeneXpert® system must be available on site. In decentralised settings with a low case-load, dried blood spot (DBS) testing might be an alternative.
METHODS: Between December 2019 and January 2021, patients with an indication for HCV RNA quantification and informed consent provided 100 µl capillary whole blood each for on-site Xpert® HCV VL FS testing (reference) and DBS testing in the laboratory. For the latter, 100 µl blood, collected with an EDTA Minivette®, were transferred to a Whatman® 903 filter card. After drying for at least 1 hour, the DBS sample was packed into a sealable plastic bag with desiccant and sent to the central laboratory of our hospital, where it was stored at –20°C. For HCV RNA extraction, the whole DBS was cut out with an 18-mm puncher and transferred into 1.3 ml guanidinium thiocyanate-containing buffer (provided by Cepheid®). After mixing and incubating at room temperature for 2–3 hours, 1 ml supernatant was analysed with the Xpert® HCV VL test (105 minutes) (filter paper absorbs 0.3 ml).
RESULTS: Of 109 paired samples from 67 patients, 38 (34.9%) were positive with the Xpert® HCV VL FS test. Sensitivity and specificity of DBS testing were 89.5% (34/38; 95% confidence interval [CI] 75.9–95.8%) and 97.2% (69/71; 95% CI 90.3–99.2%), respectively. The six (5.5%) discordant results (four false negative, two false positive) all were observed in samples with HCV RNA detectable below the limit of quantification after 2–8 weeks of pan-genotypic direct-acting antiviral treatment or 5 weeks after acute hepatitis C in a patient clearing HCV spontaneously. Quantifiable results (n = 30; 16 genotype 1, 7 genotype 3, 4 genotype 4, 1 genotype 1a and 3a, 2 unknown; HCV RNA range: 2.74–6.66 log IU/ml) correlated well (R2 = 0.981). On average, uncorrected DBS test results were 1.30 ± 0.14 log IU/ml lower than Xpert® HCV VL FS test results (~42 μl instead of the expected 1000 μl plasma used). Storage of DBS samples at room temperature for 7 days before freezing reduced HCV RNA by 0.29 ± 0.12 log IU/ml.
CONCLUSION: HCV RNA can reliably be quantified with the Xpert® HCV VL test in capillary dried blood spot samples. Thus, access to capillary HCV RNA quantification for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection can be extended to decentralised settings with a low case load.
Introduction
To achieve the World Health Organization (WHO) goal “Hepatitis C elimination by 2030”, 90% of chronic hepatitis C patients must be diagnosed and 80% treated [1]. With the well-tolerated pangenotypic direct-acting antivirals (DAAs) sofosbuvir/velpatasvir and glecaprevir/pibrentasvir [2–4], treatment duration decreased to 8–12 weeks, treatment success increased to 95–100% and genotyping became dispensable [5].
Since 2017, patients with chronic hepatitis C in Switzerland can be treated with DAAs irrespective of liver fibrosis stage [6, 7]. Between 1 May 2017 and 1 May 2019, hepatitis C virus (HCV) treatment uptake in the Swiss Association for the Medical Management in Substance Users (SAMMSU) cohort has increased from 62% to 80% [8]. However, only patients diagnosed by HCV antibody screening, further evaluated with HCV RNA polymerase chain reaction (PCR) testing and linked to care will eventually receive treatment. In 2017, it was estimated that of presumably 39,500 people with viraemic HCV infection in Switzerland, only 24,400 (62%) were diagnosed [9, 10].
Diagnostic gaps in the HCV cascade have been described all over the world [11–15], particularly for the high-prevalence population of people who inject drugs with often difficult venous access. Capillary tests are well accepted and potentially increase screening uptake [16, 17]. Reliable rapid HCV antibody tests using capillary whole blood or saliva are already available [18], but there is still need for optimising viraemia detection [19].
We recently showed that for patients with difficult venous access after long-term intravenous drug use, rapid point-of-care HCV RNA quantification in capillary whole blood with the Xpert® HCV Viral Load Fingerstick (VL FS) test (approved in September 2018; specifically developed for 100 µl whole blood; limit of detection [LOD] 40 IU/ml; limit of quantification [LOQ] 100 IU/ml; 60 minutes) [20] is a convenient and reliable method for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection [21]. For this, however, an expensive GeneXpert® system must be available on site, restricting this method to centralised settings. In decentralised settings with a low case-load, such as family practitioners or pharmacies, dried blood spot (DBS) testing might be an alternative [22–24]. In a recently published study with 170 participants (120 chronically HCV infected, 50 HCV antibody negative), Wlassow et al. showed that the Xpert® HCV VL test (for 1 ml plasma or serum; LOD 4 IU/ml for plasma, 6.1 IU/ml for serum; LOQ 10 IU/ml; 105 minutes) [25–27] is able to accurately detect and quantify HCV RNA regardless of the HCV genotype in 50 µl venous whole blood collected in an EDTA tube and spotted onto a filter paper card (DBS technique) [28]. Sensitivity was 100% (119/119, 95% confidence interval [CI] 96.9–100%) and specificity 90% (45/50, 95% CI 78.6–95.6%). On average, HCV RNA levels in whole blood were 1.93 ± 0.38 log IU/ml lower than in serum, which can be explained by the lower volume used (50 µl whole-blood eluted from a DBS versus 1000 µl serum).
The aim of our prospective study was to evaluate the diagnostic accuracy of HCV RNA quantification in a capillary dried blood spot (DBS) with the Xpert® HCV Viral Load test for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection using the Xpert® HCV Viral Load Fingerstick test, also with 100 µl capillary whole blood, as the reference test. To assess the feasibility of sending DBS samples by regular mail, we also investigated HCV RNA stability in DBSs stored at room temperature for 7 days.
Materials and methods
Ethical considerations
The study was approved by the cantonal ethical committee (AG/SO 2012/091; PB_2016-02058). All participants gave written informed consent.
Study design and participants/samples
For this prospective study, conducted between December 2019 and January 2021, patients were recruited from both the Infectious Diseases Outpatient Clinic of the Cantonal Hospital Aarau, a tertiary care hospital in Switzerland, and from the heroin substitution programme of the canton Aargau in Brugg, which was visited by an infectious disease specialist and a study nurse every 4 weeks with the GeneXpert® of the Infectious Diseases Outpatient Clinic in the framework of an HCV elimination programme running since September 2018. Adults with an indication for HCV RNA quantification and written informed consent provided 100 µl capillary whole blood each for Xpert® HCV VL FS testing (reference) and Xpert® HCV VL testing in DBS. To evaluate suitability of the Xpert® HCV VL test in DBS for HCV treatment monitoring, specimens were collected during treatment, at the end-of-treatment (EOT) and 12 weeks thereafter (sustained virological response [SVR]).
Capillary blood draw
After fingerstick with a blue BD Microtainer® contact-activated lancet (2.0 x 1.5 mm blade for high flow, enabling single puncture collection of 500 µl) (Becton Dickinson AG, Basel, Switzerland), the first 100 µl blood, collected with a 100 µµl EDTA Minivette® (Minivette® POCT 100 µl EDTA; Sarstedt, Nümbrecht, Germany), were transferred to the first circle of a Whatman® 903 filter card (WHA10531018, Sigma-Aldrich, Taufkirchen, Germany). Since each of the five circles of the filter card are intended for 75–80 µl blood, the blood spot extends 1–2 mm beyond the circle. In order to have a back-up sample in case of an error, another 100 µl blood was transferred with a second 100 µl EDTA Minivette® to the third circle of the filter card. One circle was left empty in between to prevent merging of the two blood spots. Finally, a third 100 µl EDTA Minivette® was completely filled and the blood directly used for on-site Xpert® HCV VL FS testing. After drying (at least 1 hour), the filter card was packed into a sealable plastic bag (WHA10548232, Sigma-Aldrich, Taufkirchen, Germany) with desiccant (WHA10548239, Sigma-Aldrich, Taufkirchen, Germany) and sent to the central laboratory of our hospital, where it was stored at –20°C. The filter cards were either sent by tube mail from within the hospital or by priority mail from outside, or personally brought to the laboratory (transport by private car or train). To calculate the time to laboratory and the time to measurement, the following times were documented: capillary blood draw, arrival of the filter card in the laboratory and HCV RNA measurement with the Xpert® HCV VL test.
Index test: HCV RNA quantification in capillary DBS samples with the Xpert® HCV Viral Load test
For HCV RNA extraction, the whole DBS (equivalent to 100 µl capillary whole-blood) was cut out with an 18-mm-puncher (disinfected with 70% ethanol between two samples to avoid cross-contamination) and transferred into 1.3 ml guanidinium thiocyanate-containing buffer provided by Cepheid®. After mixing (vortexing for 10–20 seconds) and incubating at room temperature for 2–3 hours, 1 ml supernatant was measured with the Xpert® HCV VL test (GXHCV-VL-CE-10; Cepheid, Sunnyvale, CA, USA), approved for 1 ml plasma (LOQ 10 IU/ml, LOD 4.0 IU/ml [95% CI 2.8–5.2], result within 105 min).
Assuming an average haematocrit of 45%, 100 µl whole blood contains ~55 µl plasma. Since only 77% (1 ml / 1.3 ml) of the added volume is transferred into the Xpert® HCV VL cartridge (0.3 ml absorbed by the filter paper), the effectively measured plasma volume is further reduced to ~42 µl which is ~24-fold less than the 1 ml for which the cartridge is approved. Thus, DBS results were expected to be 1.37 log IU/ml lower than for 1 ml plasma (correction formula: –log10(55 µl / 1000 µl × 1 ml / 1.3ml) = –log10 (0.055 × 0.77) = +1.37 log IU/ml). Xpert® HCV VL testing was done on a GeneXpert® with four modules (GXIV-4-L System; Cepheid Sunnyvale, CA, USA) in the central laboratory.
Reference test: Xpert® HCV VL FS test
One hundred microlitres capillary whole blood collected with a 100 µl EDTA Minivette® after a fingerstick were immediately transferred into an Xpert® HCV VL FS cartridge (GXHCV-FS-CE-10; Cepheid, Sunnyvale, CA, USA) (approved for 100 µl capillary whole blood; LOQ 100 IU/ml, LOD 40 IU/ml; result within 60 minutes) and measured in another GeneXpert® with four modules, which was situated either in the Infectious Diseases Outpatient Clinic or the heroin substitution programme (point-of-care-test [POCT]). A more detailed description can be found in a previous publication [21].
Stability analysis
To explore how delayed freezing due to longer transport by mail (e.g., at Easter) would affect HCV RNA levels in a DBS, two separate filter cards were collected from selected patients. Each was separately packed into a sealable plastic bag with desiccant. Upon arrival in the laboratory, one filter card was immediately stored at –20°C (within 48 h after the capillary blood draw), and the other was kept at room temperature (21°C) for 7 days prior to freezing. In one case, the second filter card remained at room temperature for 9 days and in another even for 12 days. The two differently stored DBS samples of each patient were measured in parallel and the decrease in HCV RNA level was calculated.
HCV genotyping
HCV genotypes were analysed with the VERSANT® HCV Genotype 2.0 Assay (LiPA) (Siemens, Zurich, Switzerland), detecting 6 genotypes and 19 subtypes.
Statistical analysis
The following measures of diagnostic accuracy were assessed: sensitivity (proportion of HCV RNA positive samples correctly identified by the Xpert® HCV VL test in a DBS), specificity (proportion of HCV RNA negative samples correctly identified by the Xpert® HCV VL test in a DBS), positive predictive value (proportion of individuals with a positive Xpert® HCV VL test in a DBS actually being HCV RNA positive), negative predictive value (proportion of individuals with a negative Xpert® HCV VL test in a DBS actually being HCV RNA negative) and the proportion with concordant qualitative results. Cure of chronic hepatitis C is currently defined as “HCV RNA not detectable 12 weeks after the end of treatment” (SVR12). Thus, for the sensitivity/specificity analysis, we considered only qualitative results (detectable or not). “Detectable below 100 IU/ml” (Xpert® HCV VL FS test) and “Detectable below 10 IU/ml” (Xpert® HCV VL test) counted as positive results. We also determined diagnostic accuracy according to the following indications: pre-treatment baseline, on-treatment follow-up, EOT, SVR, screening for reinfection, differentiation between spontaneous clearance and chronic hepatitis C, screening for acute infection.
Paired samples with quantifiable results in both tests underwent correlation analysis. A squared correlation coefficient (R2) and a fitted regression line were calculated overall and stratified by HCV genotype.
For paired quantifiable results, Bland-Altman plots were produced. We plotted the difference between the Xpert® HCV VL FS and the respective Xpert® HCV VL result in DBS against their average, with and without correction for the smaller plasma volume used.
Statistical analyses were performed with Stata Version 12.0 and OpenEpi (www.openepi.com).
Results
Patient/sample characteristics
Between December 2019 and January 2021, 109 paired samples were available from 67 patients; 58% (39) of the patients and 70% (76) of the samples came from the Infectious Diseases Outpatient Clinic with 42% being HCV RNA positive. The remaining 33 paired samples, with 18% HCV RNA positive, were derived from 28 patients of the heroin substitution programme.
Patient and sample characteristics are described in table 1: 73% of the patients were male, 91% had ever used intravenous drugs, 82% were on opioid agonist therapy, all but one were HCV antibody positive and 10% co-infected with human immunodeficiency virus (HIV) and HCV. The median age was 45 years. One third of the patients had more than one paired sample (up to five per patient). HCV genotype 1 was the most frequent, followed by genotype 3 and 4. On average, same-day haematocrit was 2.8% higher in men than in women.
Table 1Patient/sample characteristics.
|
Patients characteristics (n = 67)
|
| Male |
73.1% (49) |
| Median age (years) (IQR) |
44.7 (36.0–51.3) |
| Ever intravenous drug use |
91.0% (61) |
| On opioid agonist therapy |
82.1% (55) |
| HCV antibody positive |
98.5% (66) |
| HIV/HCV co-infected1
|
10.4% (7) |
| HIV mono-infected1
|
1.5% (1) |
| Proportion of patients with: |
| – One test |
65.7% (44) |
| – Two tests |
17.9% (12) |
| – Three tests |
6.0% (4) |
| – Four tests |
9.0% (6) |
| – Five tests |
1.5% (1) |
|
Sample characteristics (n = 109)
|
| HCV RNA positive with Xpert® HCV VL FS test |
34.9% (38) |
| HCV genotype distribution among the HCV RNA positives (n = 38): |
| – HCV genotype 1 |
50.0% (19) |
| – HCV genotype 3 |
23.7% (9) |
| – HCV genotype 4 |
15.8% (6) |
| – HCV genotype 1 and 3 |
2.6% (1) |
| – HCV genotype unknown |
7.9% (3) |
| Mean same-day haematocrit (%) (± SD) |
41.6 ± 4.4 (n = 39) |
| – Men |
42.9 ± 3.1 (n = 21) |
| – Women |
40.1 ± 5.2 (n = 18) |
| – Mean difference (men – women) |
2.8 (95% CI 0.1–5.5) |
Qualitative performance / diagnostic accuracy
With the Xpert® HCV VL FS test using 100 µl capillary whole blood (reference test), 34.9% (95% CI 26.6–44.2%) of the samples were HCV RNA positive. Compared with this reference test, HCV RNA detection with the Xpert® HCV VL test in a DBS of 100 µl capillary whole blood had a sensitivity of 89.5% (95% CI 75.9–95.8%) and a specificity of 97.2% (95% CI 90.3–99.2%), a positive predictive value of 94.4% (95% CI 81.9–98.5%) and a negative predictive value of 94.5% (95% CI 86.7–97.9%). The proportion with concordant qualitative results was 94.5% (95% CI 88.5–97.5%) (tables 2 and 3).
Table 2Qualitative performance / diagnostic accuracy of the Xpert® HCV Viral Load (VL) test in capillary dried blood spots (100 µl capillary whole blood).
|
Xpert® HCV VL FS, (100 µl capillary whole blood) (Reference test) |
| Detected |
Not detected |
Total |
| Xpert® HCV VL in capillary dried blood spot, (100 µl capillary whole blood) |
Detected |
34 |
2a
|
36 |
| Not detected |
4b
|
69 |
73 |
| Total |
38 |
71 |
109 |
For all four false negative results, the Xpert® HCV VL FS result was “HCV detected <100 IU/ml”. Three samples had been collected 2–4 weeks after DAA treatment initiation and one 5 weeks after the diagnosis of an acute hepatitis C in a patient with spontaneous clearance.
For both false positive results collected 4 and 8 weeks after DAA treatment initiation, the Xpert® HCV VL result in DBS was “HCV detected <10 IU/ml”. In the first patient, the undetectable Xpert® HCV VL FS result was confirmed by a Cobas® test in 650 µl venous EDTA plasma from the same day.
Quantitative performance
Thirty samples (53.3%, n = 16, genotype 1; 23.3%, n = 7 genotype 3; 13.3%, n = 4, genotype 4; 3.3%, n = 1, genotype 1a and 3a; 6.7%, n = 2, genotype unknown) with quantifiable results in both tests underwent correlation analysis. The HCV RNA range with the Xpert® HCV VL FS test was 553–4,540,000 IU/ml, 2.74–6.66 log IU/ml; the HCV range with the Xpert® HCV VL test in DBS was 30–344,000 IU/ml, 1.48–5.54 log IU/ml, for the raw data and 708–8,128,305 IU/ml, 2.85–6.91 log IU/ml, after correction for the smaller plasma volume.
Xpert® HCV VL FS results and Xpert® HCV VL results in DBS were highly correlated (R2 = 0.9811) irrespective of HCV-genotype (fig. 1). The equation for the fitted regression line was:
Xpert® HCV VL in DBS (corrected for the smaller plasma volume) (log IU/ml) = 1.0314 × Xpert® HCV VL FS (log IU/ml) – 0.0996
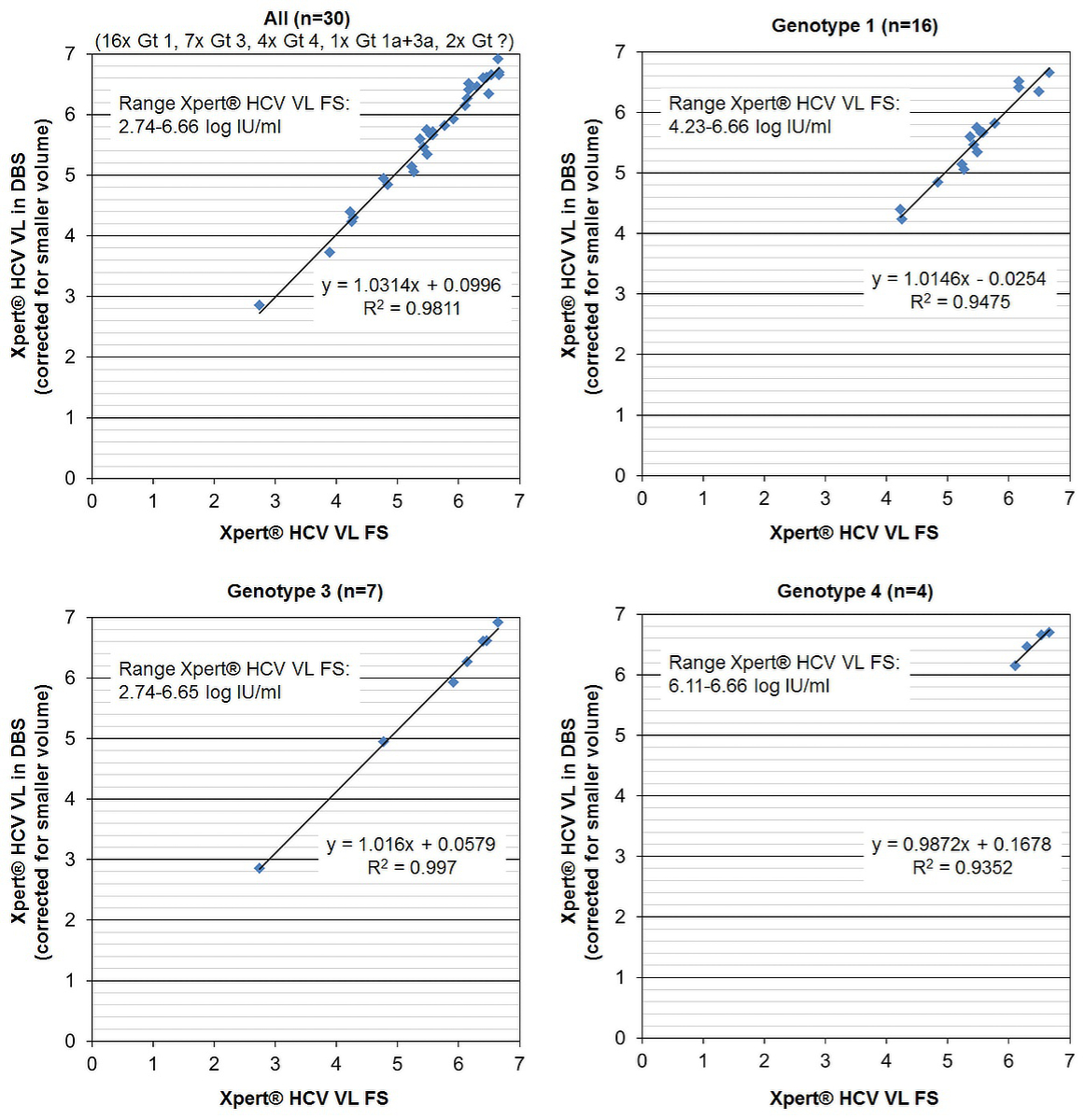
Figure 1 Quantitative performance of the Xpert® HCV Viral Load (VL) test in dried blood spot (DBS) produced with 100 µl capillary whole blood. Correlation of quantifiable HCV RNA results (log IU/ml) with the reference test Xpert® HCV Viral Load Fingerstick (VL FS), overall and according to genotype (Gt).
On average, uncorrected Xpert® HCV VL results in DBS were 1.30 ± 0.14 log IU/ml lower (range 1.03–1.59) than the corresponding Xpert® HCV VL FS result. The Bland-Altman plot (supplementary fig. S1 in the appendix) showed an average bias of 1.30 (95% CI 1.25–1.35). The 95% limits of agreement were 1.02 to 1.58. After correction for the smaller plasma volume, corrected Xpert® HCV VL results in DBS were on average 0.07 ± 0.14 log IU/ml higher (range –0.22 to 0.34) than the corresponding Xpert® HCV VL FS result. The Bland-Altman plot (supplementary fig. S2) showed an average bias of –0.08 (95% CI –0.13 to –0.02). The 95% limits of agreement were –0.35 to 0.20.
Detectable below the LOQ
Seven Xpert® HCV VL FS results were “detectable below the LOQ” (100 IU/ml, 2.0 log IU/ml). Six of the seven samples were collected 2–4 weeks after DAA treatment initiation and one 5 weeks after the diagnosis of acute hepatitis C in a patient with spontaneous clearance. Four were undetectable with the Xpert® HCV VL-test in DBS (false negative) and three “detectable below the LOQ” (10 IU/ml, 1.0 log IU/ml).
On the other hand, six Xpert® HCV VL tests in DBS were “detectable below the LOQ” (10 IU/ml, 1.0 log IU/ml). All six samples were collected 2–8 weeks after DAA treatment initiation. Two were undetectable with the Xpert® HCV VL FS test (false positive), three were “detectable below the LOQ” (100 IU/ml, 2.0 log IU/ml) and one had a quantifiable result, i.e. 157 IU/ml = 2.2 log IU/ml.
Performance according to indication
Table 3 shows test performance according to HCV RNA indication. Seventy-three percent (80/109) of all tests were off-treatment (pre-treatment baseline, SVR, screening for acute infection/reinfection, differentiation between spontaneous clearance [SC] and chronic hepatitis C). Off-treatment, the only discrepancy was a false negative result in a patient with spontaneous clearance 5 weeks after the diagnosis of an acute hepatitis C. Accordingly, concordance, sensitivity and specificity were 98.8% (79/80, 95% CI 93.3–99.8%), 96.7% (29/30, 95% CI 83.3–99.4%) and 100% (50/50, 95% CI 92.9–100%), respectively. With a prevalence of 37.5% (30/80), positive and negative predictive values were 100% (29/29, 95% CI 88.3–100%) and 98.0% (50/51, 95% CI 89.7–99.7%), respectively.
Table 3Performance according to indication (Xpert® HCV Viral Load (VL) test in dried blood spots).
|
Xpert® HCV VL-test in DBS produced with 100 µl capillary whole blood (n = 109)
|
|
|
Indication
|
% of tests (n)
|
Prevalence (%)
|
Concordance (%)
|
Sensitivity (%)
|
Specificity (%)
|
PPV (%)
|
NPV (%)
|
| Pre-treatment baseline |
21.1 (23) |
100 (23/23) |
100 (23/23) |
100 (23/23) |
– |
100 (23/23) |
– |
| On-treatment follow-up (weeks 1–8) |
15.6 (17) |
47.1 (8/17) |
70.6 (12/17) |
62.5 (5/8) |
77.8 (7/9) |
71.4 (5/7) |
70.0 (7/10) |
| EOT |
11.0 (12) |
0 (0/12) |
100 (12/12) |
– |
100 (12/12) |
– |
100 (12/12) |
| SVR |
12.8 (14) |
0 (0/14) |
100 (14/14) |
– |
100 (14/14) |
– |
100 (14/14) |
| Reinfection? |
30.3 (33) |
0 (0/33) |
100 (33/33) |
– |
100 (33/33) |
– |
100 (33/33) |
| SC or chronic hepatitis C? |
8.3 (9) |
77.8 (7/9) |
88.9 (8/9) |
85.7 (6/7) |
100 (2/2) |
100 (6/6) |
66.7 (2/3) |
| Acute infection? |
1.0 (1) |
0 (0/1) |
100 (1/1) |
– |
100 (1/1) |
– |
100 (1/1) |
| All |
100 (109) |
34.9 (38/109) |
94.5 (103/109) |
89.5 (34/38) |
97.2 (69/71) |
94.4 (34/36) |
94.5 (69/73) |
Twenty-seven percent (29/109) of all tests were on-treatment (on-treatment follow-up [weeks 1–8] or EOT). Due to three false negative Xpert® HCV VL tests in DBS samples collected in treatment weeks 2, 3 and 4, sensitivity and negative predictive value were reduced to 62.5% (5/8, 95% CI 30.6–86.3%) and 86.4% (19/22, 95% CI 66.7–95.3%), respectively. Due to two false positive Xpert® HCV VL tests in DBS collected in treatment weeks 4 and 8, specificity and positive predictive value declined to 90.5% (19/21, 95% CI,: 71.1–97.4) and 71.4% (5/7, 95% CI 35.9–91.8), respectively.
HCV RNA monitoring prior to and during DAA treatment
Figure 2 shows the HCV RNA course in eight exemplary patients. HCV RNA levels were measured in parallel with the Xpert® HCV VL FS test and the Xpert® HCV VL test in DBS samples prior to, during and after DAA treatment as well as during spontaneous clearance. Additionally, available HCV RNA measurements with the Cobas® assay are also displayed.
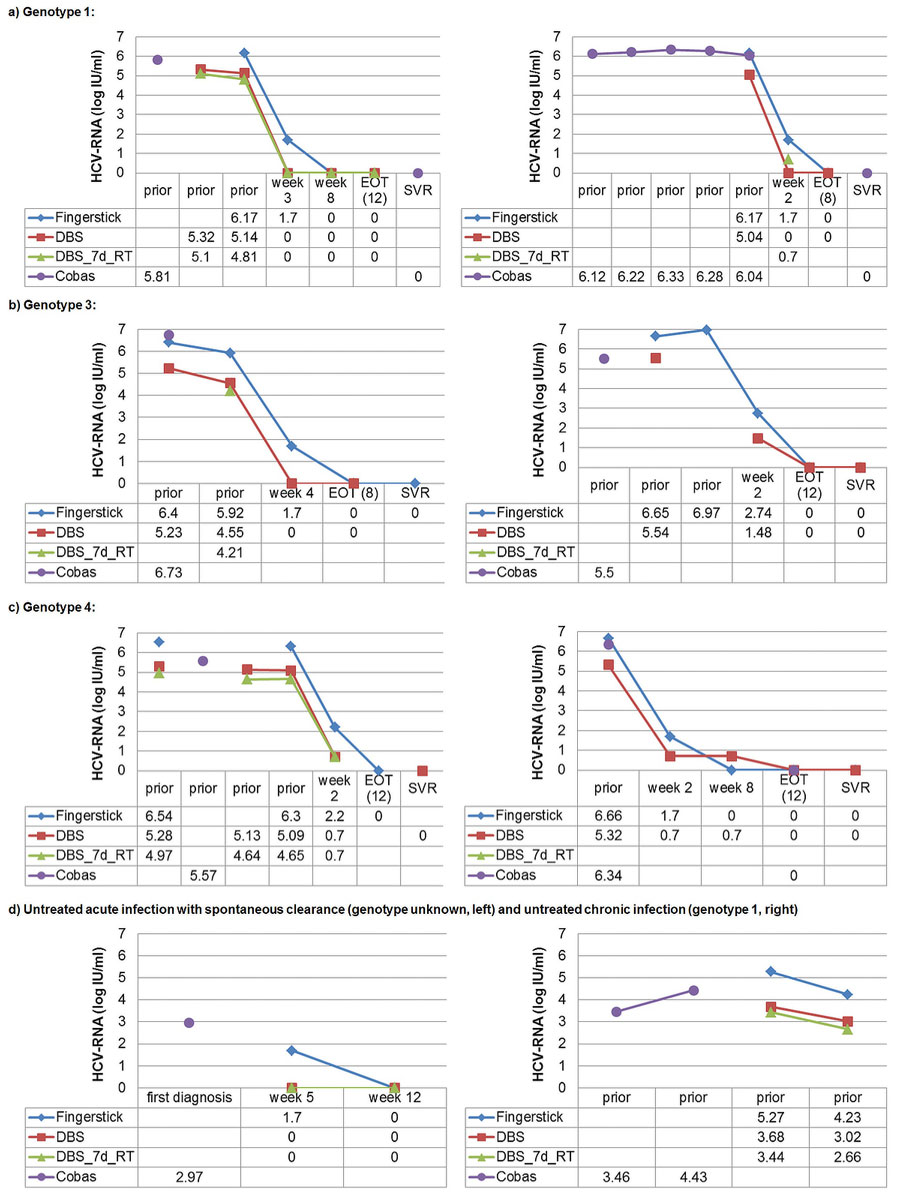
Figure 2 HCV RNA course in eight exemplary patients with different genotypes: prior to, during and after direct-acting antiviral (DAA) treatment as well as during untreated acute and chronic infection.
Fingerstick = Xpert® HCV Viral Load Fingerstick-test (limit of quantification 100 IU/ml, 2.0 log IU/ml); DBS = Xpert® HCV Viral Load test (limit of quantification 10 IU/ml, 1.0 log IU/ml) in DBS samples produced with 100 µl capillary whole-blood and stored at –20°C within 48 hours after capillary blood draw (uncorrected result); DBS_7d-RT = Xpert® HCV Viral Load test in DBS samples produced with 100 µl capillary whole blood and exposed to room temperature for 7 days prior to storage at –20°C (uncorrected result); Cobas = Cobas® assay with 650 µl venous plasma (limit of quantification 15 IU/ml, 1.2 log IU/ml); prior = prior to HCV treatment; week 2, week 3, week 4, week 8 = respective weeks under HCV treatment; EOT (8) = end of treatment (week 8); EOT (12) = end of treatment (week 12); SVR = sustained virological response; 0 = HCV RNA not detectable; first diagnosis = first diagnosis of acute hepatitis C; week 5, week 12 = respective week after diagnosis of acute hepatitis C.
For unquantifiable positive Xpert® HCV Viral Load Fingerstick test results (i.e., “detectable below the limit of quantification of 100 IU/ml, 2.0 log IU/ml”), the mid-point between zero and the lower limit of quantification was used (i.e., 50 IU/ml, 1.70 log IU/ml). For unquantifiable positive Xpert® HCV Viral Load test results (i.e., “detectable below the limit of quantification of 10 IU/ml, 1.0 log IU/ml”), the mid-point between zero and the lower limit of quantification was used (i.e., 5 IU/ml, 0.70 log IU/ml).
Irrespective of genotype and DAA regimen used, there was a sharp HCV RNA decline during the first 2–4 weeks of treatment towards “undetectable” or a value near the limit of quantification (fig. 2a–c). The same was true in a patient with spontaneous clearance 5 weeks after the diagnosis of an acute hepatitis C (fig 2d).
In most patients, the HCV RNA level was relatively stable prior to treatment (fig. 2a–c). In one patient with an HCV RNA fluctuation of ≥1 log IU/ml, the Xpert® HCV VL FS test, the Xpert® HCV VL test in a DBS stored at –20°C within 48 hours after the capillary blood draw and the Xpert® HCV VL test in a DBS exposed to room temperature for 7 days prior to storage at –20°C showed a parallel course (fig. 2d).
HCV RNA stability in DBS samples at room temperature
To assess HCV RNA stability, 33 paired DBS samples of 19 different patients were analysed (up to 5 paired samples/patient). Of each sample pair, one was stored at –20°C within 48 hours, and the other remained at room temperature for 7 (n = 31), 9 (n = 1) or 12 days (n = 1) prior to storage at 20°C. After 7 days at room temperature, the average HCV RNA decrease was 0.29 ± 0.12 log IU/ml (range 0.05–0.49; n = 18). It was similar for genotype 1 (0.30 ± 0.11 log IU/ml, range 0.16–0.47; n = 10), genotype 3 (0.29 ± 0.14 log IU/ml, range 0.08–0.40; n = 4) and genotype 4 (0.28 ± 0.22, range 0.06–0.49; n = 3). In the patient with genotypes 1a and 3a, the HCV RNA decrease was 0.29 log IU/ml. Even after 9 and 12 days at room temperature, the HCV RNA decrease was less than 0.5 log U/ml: 0.44 log IU/ml (genotype 4) and 0.46 log IU/ml (genotype 1), respectively (fig. 3).
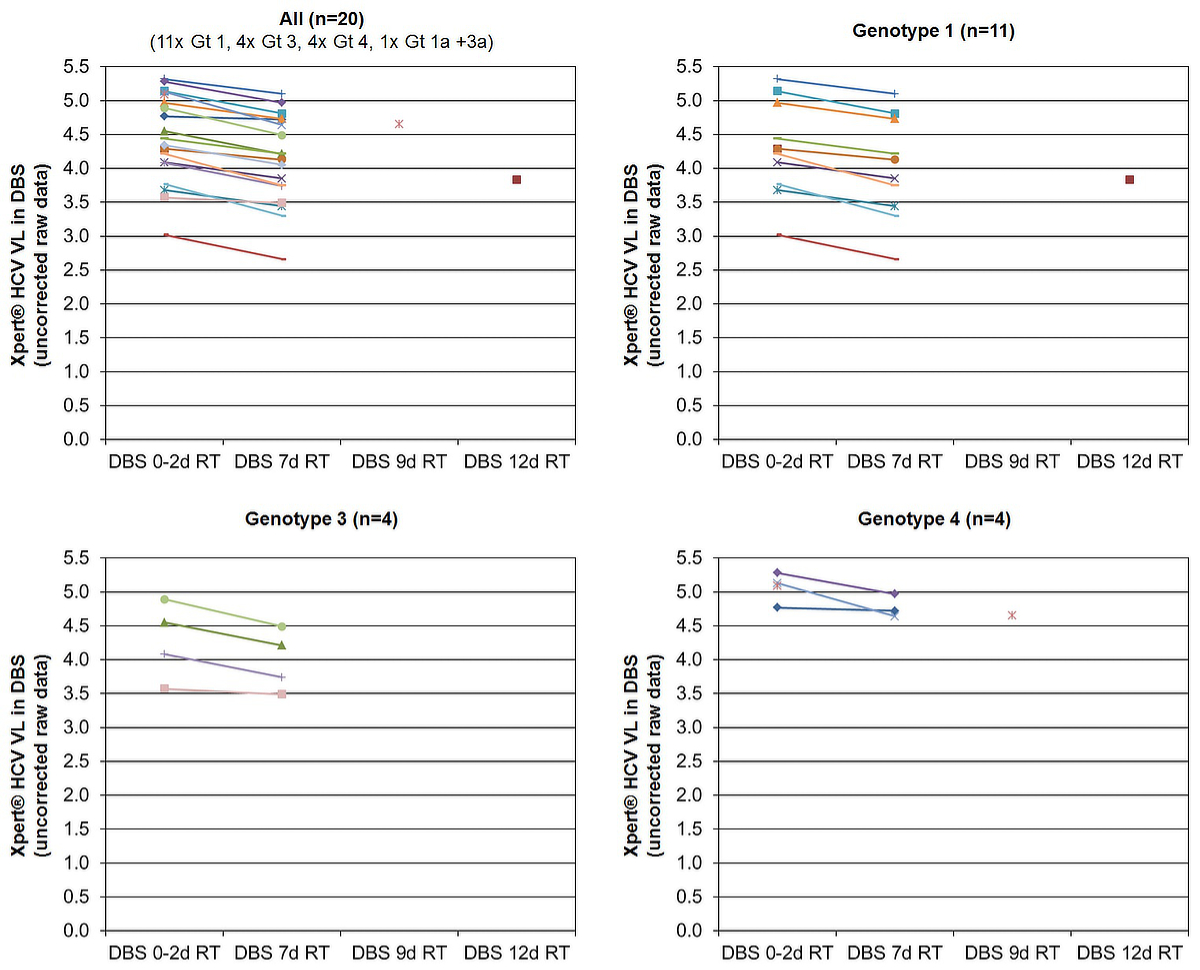
Figure 3 HCV RNA stability in dried blood spot samples (DBS) exposed to room temperature for 7 days prior to storage at 20°C
Gt = genotype; Xpert® HCV VL = Xpert® HCV Viral Load test; RT = room temperature; DBS 0–2d RT = DBS stored at –20°C within 48 hours after capillary blood draw; DBS 7d RT = DBS exposed to room temperature for 7 days prior to storage at –20°C; DBS 9d RT = DBS exposed to room temperature for 9 days prior to storage at –20°C; DBS 12d RT = DBS exposed to room temperature for 12 days prior to storage at –20°C
For 29 of the 33 paired DBS samples, a corresponding Xpert® HCV VL FS result was available. All of the 18 quantifiable HCV RNA positive samples (HCV RNA range with the Xpert® HCV VL FS test: 157–3,440,000 IU/ml, 2.2–6.54 log IU/ml) were detected with the Xpert® HCV VL test in the DBS sample regardless of whether they were immediately (within 48 hours) stored at 20°C or initially stored at room temperature for 7–12 days. Of the five samples that were “detected <100 IU/ml” with the Xpert® HCV VL FS test (week 2–4 under DAA treatment and week 5 after diagnosis of an acute hepatitis C in a patient with spontaneous clearance), three were “detected <10 IU/ml” with the Xpert® HCV VL test in the corresponding DBS sample stored for 7 days at room temperature. For all six samples in which no HCV RNA was detected with the Xpert® HCV VL FS test, the Xpert® HCV VL test in the corresponding DBS sample stored for 7 days at room temperature also yielded “HCV not detected”. Thus, even if the DBS sample remained 7–12 days at room temperature prior to storage at 20°C, sensitivity and specificity were still 91.3% (21/23) and 100% (6/6), respectively. If only off-treatment samples were considered, sensitivity was 94.4% (17/18).
Time to laboratory and time to measurement
Of the 109 paired samples, 74 (67.9%) filter cards were transported within the hospital by tube mail. These samples, all coming from the Infectious Diseases Outpatient Clinic, arrived in the laboratory within 24 hours after the capillary blood draw (median time to lab 4.8 hours, IQR 3–6.3 hours, range: 0–21 hours; n = 68).
Twenty-two samples (20.2%) were personally brought to the laboratory. All but two came from the heroin substitution programme of the canton Aargau and were transported either by private car or train. Except for one sample (blood draw on Friday, transport on Monday), all samples arrived in the laboratory within 48 hours after the blood draw (median time to lab 4.7 hours, IQR 3–7.4 hours, range: 2.3–64.4 hours; n = 22).
Thirteen samples (11.9%), coming from the heroin substitution programme of the canton Aargau, were sent by priority mail. All but two arrived in the laboratory within 48 hours (median time to lab 44 hours, IQR 26.1–45 hours range 18–64.5 hours; n = 13).
Of the DBS samples, 75.2% (82/109) were measured with the Xpert® HCV VL test within 14 days after the blood draw (median time to measurement 8.9 days, IQR 7–13.9, range 1–28.2 days; n = 109). Supply shortage of the Xpert® HCV VL test due to the COVID19-pandemic resulted in delayed measurements.
Discussion
Main findings
HCV RNA can reliably be detected and quantified with the Xpert® HCV VL test in DBS samples from 100 µl capillary whole blood. Discordant results compared with the Xpert® HCV VL FS test only occurred under antiviral treatment or spontaneous clearance with HCV RNA levels detectable below the limit of quantification, and were thus probably not clinically relevant. Quantifiable results correlated well, irrespective of genotype. Since only ~42 µl instead of 1000 µl plasma were used, uncorrected Xpert® HCV VL test results in DBS were on average 1.3 log IU/ml lower than the corresponding Xpert® HCV VL FS result. Storage of DBS samples at room temperature for 7 days reduced HCV RNA by <0.3 log IU/ml, allowing transport by regular mail.
Diagnostic accuracy
In a meta-analysis of 15 studies evaluating the diagnostic accuracy of HCV RNA in DBS, both pooled sensitivity and specificity were 98% (95% CI 95–99) [23]. In the proof-of-principle study of Wlassow et al. in 120 chronically HCV infected and 50 HCV negative patients, HCV RNA quantification with the Xpert® HCV VL test in DBS samples from 50 µl venous whole blood showed a sensitivity and specificity of 100% (95% CI 96.9–100) and 90.0% (95% CI 78.6–95.6), respectively, compared with the Xpert® HCV VL test with 1000 µl serum [28]. False positive results might result from cell-associated HCV RNA in B lymphocytes present in whole blood but not in plasma/serum [22, 29]. Consistent with this hypothesis, Stapleton et al. found that HCV RNA detection in whole blood was more sensitive than plasma-based methods [22, 30]. In our study, using 100 µl whole blood each for index and reference test and including 27% samples under DAA treatment, sensitivity was only 89.5% (34/38, 95% CI 75.9–95.8) and specificity 97.2% (69/71, 95% CI 90.3–99.2). However, all six discordant results were under pan-genotypic DAA treatment or spontaneous clearance with HCV RNA detectable below the limit of quantification, and thus probably not clinically relevant. Restricting the analysis to the 80 paired samples collected off-treatment, the only remaining discrepancy was a false negative result in a patient undergoing spontaneous clearance 5 weeks after acute hepatitis C. Accordingly, sensitivity and specificity increased to 96.7% (29/30) and 100% (50/50), respectively, which is in line with the meta-analysis by Lange et al. [23].
In 2010, Tuaillon et al. observed 2.27 ± 0.47 log IU/ml lower viral loads in DBSs [31]. In 2016, Soulier et al. reported 1.60 ± 0.3 log IU/ml and 1.75 ± 0.3 log IU/ml lower results [32], depending on the HCV RNA assay used. In our analysis, we show only 1.3 ± 0.14 log IU/ml lower results in DBSs, which may be explained by improved HCV RNA extraction methods and twice as much blood (100 µl instead of 50 µl) for the DBS sample corresponding to 0.3 log UI/ml higher values.
HCV RNA stability in DBS samples at room temperature
Whereas HIV RNA and hepatitis B virus DNA are considered to be stable in DBS samples at room temperature [23, 33], this is less clear for HCV RNA. Tuaillon et al. found a 3-fold decrease (–0.48 log IU/ml) in HCV RNA levels after 6 days at room temperature compared with those stored at 20°C [31]. Another study, using dried serum spots, even showed a 10-fold reduction (1 log IU/ml) of virus yield after 4 weeks compared with frozen samples, yet without reduction of the positivity rate [34].
In our study, after 7 days at room temperature, the average HCV RNA decrease was <0.3 log IU/ml (n = 18), with no difference among genotypes. After 9 and 12 days (one sample each), the HCV RNA decrease was still <0.5 log IU/ml. This is consistent with Soulier et al., who observed a HCV RNA reduction of 0.2 ± 0.15 log IU/ml between paired samples stored at 80°C and room temperature (24°C) for 19 ± 1 months [32]. Similarly, Catlett et al. found a maximum of 0.1 log IU/ml degradation after 60 days [35]. Two other studies did not find a decrease in HCV RNA after prolonged (>11 months) periods at room temperature [24, 36]. Thus, sending DBS samples for HCV RNA quantification by regular mail seems unproblematic.
Clinical implication and future research
On the basis of what we actually need for diagnosing chronic HCV infection, monitoring DAA treatment and detecting reinfection, the performance of the Xpert® HCV VL test in a DBS exceeds the needs. According to current guidelines [37–39], HCV RNA monitoring under DAA treatment, i.e. at week 2–4 (adherence assessment by documenting an HCV RNA decrease) and at EOT, is optional. At diagnosis/baseline and SVR check, patients are off-treatment, for which sensitivity and specificity of the Xpert® HCV VL test in a DBS were 97% and 100%, respectively. Since >95% of treatment-naïve chronic hepatitis C patients have a viral load >10,000 (>4 log) IU/ml [40–42], a test with a detection threshold of 1000 (3 log) IU/ml is still acceptable [38, 42].
In drug services in Scotland, the introduction of DBS sampling resulted in a 3-fold increase in testing and a 12-fold increase in positives [17]. In the decentralised opioid agonist therapy (OAT) setting of the canton Aargau [12], with >80% of opioid agonists dispensed in pharmacies (www.substitution.ch) [43], the logical next step is to offer HCV screening and treatment in pharmacies [44–50]. Patients with unknown HCV serostatus or a negative HCV antibody test >1 year ago [51] should undergo an HCV antibody rapid test from either saliva or capillary whole blood (OraQuick®, result within 5–20 min) [52–56]. If positive, a capillary DBS sample should immediately be collected for HCV RNA quantification with the Xpert® HCV VL test in the laboratory. Similarly, HCV antibody positive patients with unknown HCV RNA status or a negative HCV RNA test >1 year ago [51] should directly have capillary DBS sampling for HCV RNA testing.
In contrast to Australia [57–59] and France [60], general practitioners in Switzerland are not allowed to prescribe DAA treatment. Referral to a specialist (gastroenterologist or infectious disease specialist) is often unrewarding because OAT patients often have difficulties keeping appointments [11, 61]. Therefore, HCV RNA positive patients can be linked to care/treatment via www.hep-care.ch, allowing DAA prescription by a specialist based only on the patient’s medical records. DAAs can then be dispensed together with the OAT in the pharmacy and HCV RNA monitored with DBS samples.
To facilitate widespread clinical use of the Xpert® HCV VL test for HCV RNA quantification in DBS, Cepheid® should seek regulatory approval for a DBS package insert claim [35], as already exists for the Xpert® HIV VL test [62].
Strengths and limitations of the study
Our study has several strengths. Whereas others pipetted remnant venous whole blood onto a filter card in the laboratory [28, 32], we prospectively collected fingerstick capillary whole blood directly onto filter paper, which reflects the “real world”. Using a completely filled 100 µl EDTA Minivette® and cutting out the whole DBS with an 18 mm puncher, we have probably analysed a higher volume than others using only 50 µl blood and a 6 mm or 12 mm puncher [28, 31, 32]. Correlation between Xpert® HCV VL in DBS and Xpert® HCV VL FS as well as HCV RNA stability in DBS at room temperature were analysed stratified by genotype. Unfortunately, there were only a few samples with genotype 4 and no samples with genotypes 2, 5 and 6. Patients under DAA treatment were not excluded, but deliberately monitored throughout their HCV treatment up to the SVR check-up. Since both our index and reference tests were performed from 100 µl capillary whole-blood, we could easily recruit patients with difficult venous access from the heroin substitution programme for our study. As a consequence of the 100 µl blood volume, the LOD (40 IU/ml) and LOQ (100 IU/ml) of our reference test were higher than in other studies using Cobas® (15 IU/ml), Abbott® (12 IU/ml) or Xpert® (4 IU/ml; 10 IU/ml) in 650 µl, 500 µl and 1000 µl venous plasma/serum, respectively. Another limitation is the relatively low number of HCV RNA positive samples. As screening for reinfection and acute infection as well as HCV RNA determination at the EOT- and SVR checks did not yield positive results, we could not calculate sensitivity and positive predictive value for these indications.
Conclusion
HCV RNA can reliably be quantified with the Xpert® HCV VL test in capillary DBS samples. Thus, access to capillary HCV RNA quantification for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection can be extended to decentralised settings with a low case load such as family practitioners or pharmacies. HCV RNA stability in DBS samples for at least 7 days at room temperature allows transport by regular mail.
Acknowledgement
We thank Petra Rippstein, Stefanie Wendel and Martin Ooms from the heroin substitution programme of the canton Aargau in Brugg for their support.
Preliminary results were presented as a poster at the Annual Meeting of the Swiss Society for Infectious Diseases in Geneva, September 2–4, 2020.
Andrea Bregenzer, MD, MSc
Department of Infectious Diseases and Hospital Hygiene
Cantonal Hospital Aarau
Tellstrasse 25
CH-5001 Aarau
andrea.bregenzer[at]ksa.ch
References
1.
WHO
. 05/2016, Combating hepatitis B and C to reach elimination by 2030 - Advocacy brief; http://apps.who.int/iris/bitstream/10665/206453/1/WHO_HIV_2016.04_eng.pdf (Accessed: 2121 May 21)
2.
Chahine EB
,
Sucher AJ
,
Hemstreet BA
. Sofosbuvir/Velpatasvir: The First Pangenotypic Direct-Acting Antiviral Combination for Hepatitis C. Ann Pharmacother. 2017 Jan;51(1):44–53. https://doi.org/10.1177/1060028016668897
3.
Puoti M
,
Foster GR
,
Wang S
,
Mutimer D
,
Gane E
,
Moreno C
, et al.
High SVR12 with 8-week and 12-week glecaprevir/pibrentasvir therapy: an integrated analysis of HCV genotype 1-6 patients without cirrhosis. J Hepatol. 2018 Aug;69(2):293–300. https://doi.org/10.1016/j.jhep.2018.03.007
4.
Brown RS Jr
,
Buti M
,
Rodrigues L
,
Chulanov V
,
Chuang WL
,
Aguilar H
, et al.
Glecaprevir/pibrentasvir for 8 weeks in treatment-naïve patients with chronic HCV genotypes 1-6 and compensated cirrhosis: the EXPEDITION-8 trial. J Hepatol. 2020 Mar;72(3):441–9. https://doi.org/10.1016/j.jhep.2019.10.020
5.
Aghemo A
,
Colombo M
. Glecaprevir/Pibrentasvir: The Final Piece in the Hepatitis C Virus Treatment Puzzle? Gastroenterology. 2018 Mar;154(4):1195–6. https://doi.org/10.1053/j.gastro.2018.01.035
6.
Bundesamt für Gesundheit (BAG)
. BAG erweitert Vergütung von Medikamenten gegen Hepatitis C (27 Apr 17). Available from: https://www.bag.admin.ch/bag/de/home/das-bag/aktuell/medienmitteilungen.msg-id-66508.html
7.
Bundesamt für Gesundheit (BAG)
. Hepatitis C: Uneingeschränkte Vergütung der neuen Arzneimittel für alle Betroffenen (25 Sept 2017). Available from: https://www.bag.admin.ch/bag/de/home/das-bag/aktuell/medienmitteilungen.msg-id-68158.html
8.
Bregenzer A
,
Bruggmann P
,
Castro E
,
Moriggia A
,
Rothen M
,
Thurnheer MC
, et al.
Hepatitis C virus elimination in Swiss opioid agonist therapy programmes - the SAMMSU cohort. Swiss Med Wkly. 2021 Mar;151:w20460. https://doi.org/10.4414/smw.2021.20460
9. Swiss Hepatitis Strategy 2014-2030 (January 2019, Version 4). Available from: https://www.hepatitis-schweiz.ch/download/2871/Process_Paper_14_02_2019.pdf (Accesssed: 22.02.21)
10.
Zahnd C
,
Brezzi M
,
Bertisch B
,
Giudici F
,
Keiser O
. Analyse de Situation des Hépatites B et C en Suisse. Available from: https://www.bag.admin.ch/dam/bag/de/dokumente/mt/forschungsberichte/situationsanalyse-hepatitis-zusammenfassung.pdf.download.pdf/situationsanalyse-hepatitis-zusammenfassung-de.pdf (Accessed: 22.05.21)
11.
Schürch S
,
Fux CA
,
Dehler S
,
Conen A
,
Knuchel J
,
Friedl A
, et al.
Management of hepatitis C in opioid agonist therapy patients of the Swiss canton Aargau within and outside the cohort study. Swiss Med Wkly. 2020 Aug;150:w20317. https://doi.org/10.4414/smw.2020.20317
12.
Bregenzer A
,
Conen A
,
Knuchel J
,
Friedl A
,
Eigenmann F
,
Näf M
, et al.
Management of hepatitis C in decentralised versus centralised drug substitution programmes and minimally invasive point-of-care tests to close gaps in the HCV cascade. Swiss Med Wkly. 2017 Nov;147:w14544.
13.
Janjua NZ
,
Kuo M
,
Yu A
,
Alvarez M
,
Wong S
,
Cook D
, et al.
The Population Level Cascade of Care for Hepatitis C in British Columbia, Canada: The BC Hepatitis Testers Cohort (BC-HTC). EBioMedicine. 2016 Oct;12:189–95. https://doi.org/10.1016/j.ebiom.2016.08.035
14.
Yehia BR
,
Schranz AJ
,
Umscheid CA
,
Lo Re V 3rd
. The treatment cascade for chronic hepatitis C virus infection in the United States: a systematic review and meta-analysis. PLoS One. 2014 Jul;9(7):e101554. https://doi.org/10.1371/journal.pone.0101554
15.
Hajarizadeh B
,
Grebely J
,
McManus H
,
Estes C
,
Razavi H
,
Gray RT
, et al.
Chronic hepatitis C burden and care cascade in Australia in the era of interferon-based treatment. J Gastroenterol Hepatol. 2017 Jan;32(1):229–36. https://doi.org/10.1111/jgh.13453
16.
Hayes B
,
Briceno A
,
Asher A
,
Yu M
,
Evans JL
,
Hahn JA
, et al.
Preference, acceptability and implications of the rapid hepatitis C screening test among high-risk young people who inject drugs. BMC Public Health. 2014 Jun;14(1):645. https://doi.org/10.1186/1471-2458-14-645
17.
McLeod A
,
Weir A
,
Aitken C
,
Gunson R
,
Templeton K
,
Molyneaux P
, et al.
Rise in testing and diagnosis associated with Scotland’s Action Plan on Hepatitis C and introduction of dried blood spot testing. J Epidemiol Community Health. 2014 Dec;68(12):1182–8. https://doi.org/10.1136/jech-2014-204451
18.
Tang W
,
Chen W
,
Amini A
,
Boeras D
,
Falconer J
,
Kelly H
, et al.
Diagnostic accuracy of tests to detect Hepatitis C antibody: a meta-analysis and review of the literature. BMC Infect Dis. 2017 Nov;17(S1 Suppl 1):695. https://doi.org/10.1186/s12879-017-2773-2
19.
Ivanova Reipold E
,
Easterbrook P
,
Trianni A
,
Panneer N
,
Krakower D
,
Ongarello S
, et al.
Optimising diagnosis of viraemic hepatitis C infection: the development of a target product profile. BMC Infect Dis. 2017 Nov;17(S1 Suppl 1):707. https://doi.org/10.1186/s12879-017-2770-5
20.
Lamoury FM
,
Bajis S
,
Hajarizadeh B
,
Marshall AD
,
Martinello M
,
Ivanova E
, et al.; LiveRLife Study Group
. Evaluation of the Xpert HCV Viral Load Finger-Stick Point-of-Care Assay. J Infect Dis. 2018 May;217(12):1889–96. https://doi.org/10.1093/infdis/jiy114
21.
Bregenzer A
,
Warmann N
,
Ottiger C
,
Fux CA
. Rapid point-of-care HCV RNA quantification in capillary whole blood for diagnosing chronic HCV infection, monitoring treatment and detecting reinfection. Swiss Med Wkly. 2019 Oct;149:w20137. https://doi.org/10.4414/smw.2019.20137
22.
Greenman J
,
Roberts T
,
Cohn J
,
Messac L
. Dried blood spot in the genotyping, quantification and storage of HCV RNA: a systematic literature review. J Viral Hepat. 2015 Apr;22(4):353–61. https://doi.org/10.1111/jvh.12345
23.
Lange B
,
Roberts T
,
Cohn J
,
Greenman J
,
Camp J
,
Ishizaki A
, et al.
Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples - a systematic review and meta-analysis. BMC Infect Dis. 2017 Nov;17(S1 Suppl 1):693. https://doi.org/10.1186/s12879-017-2776-z
24.
Bennett S
,
Gunson RN
,
McAllister GE
,
Hutchinson SJ
,
Goldberg DJ
,
Cameron SO
, et al.
Detection of hepatitis C virus RNA in dried blood spots. J Clin Virol. 2012 Jun;54(2):106–9. https://doi.org/10.1016/j.jcv.2012.02.004
25.
McHugh MP
,
Wu AH
,
Chevaliez S
,
Pawlotsky JM
,
Hallin M
,
Templeton KE
. Multicenter Evaluation of the Cepheid Xpert Hepatitis C Virus Viral Load Assay. J Clin Microbiol. 2017 May;55(5):1550–6. https://doi.org/10.1128/JCM.02460-16
26.
Gupta E
,
Agarwala P
,
Kumar G
,
Maiwall R
,
Sarin SK
. Point -of -care testing (POCT) in molecular diagnostics: performance evaluation of GeneXpert HCV RNA test in diagnosing and monitoring of HCV infection. J Clin Virol. 2017 Mar;88:46–51. https://doi.org/10.1016/j.jcv.2017.01.006
27. Cepheid®, Home → Tests → Virology → Xpert HCV Viral Load, https://www.cepheid.com/en/tests/Virology/Xpert-HCV-Viral-Load (Accessed: 2021 May 22)
28.
Wlassow M
,
Poiteau L
,
Roudot-Thoraval F
,
Rosa I
,
Soulier A
,
Hézode C
, et al.
The new Xpert HCV viral load real-time PCR assay accurately quantifies hepatitis C virus RNA in serum and whole-blood specimens. J Clin Virol. 2019 Aug;117:80–4. https://doi.org/10.1016/j.jcv.2019.06.007
29.
Santos C
,
Reis A
,
Dos Santos CV
,
Damas C
,
Silva MH
,
Viana MV
, et al.
The use of real-time PCR to detect hepatitis C virus RNA in dried blood spots from Brazilian patients infected chronically. J Virol Methods. 2012 Jan;179(1):17–20. https://doi.org/10.1016/j.jviromet.2011.06.012
30.
Stapleton JT
,
Klinzman D
,
Schmidt WN
,
Pfaller MA
,
Wu P
,
LaBrecque DR
, et al.
Prospective comparison of whole-blood- and plasma-based hepatitis C virus RNA detection systems: improved detection using whole blood as the source of viral RNA. J Clin Microbiol. 1999 Mar;37(3):484–9. https://doi.org/10.1128/JCM.37.3.484-489.1999
31.
Tuaillon E
,
Mondain AM
,
Meroueh F
,
Ottomani L
,
Picot MC
,
Nagot N
, et al.
Dried blood spot for hepatitis C virus serology and molecular testing. Hepatology. 2010 Mar;51(3):752–8.
32.
Soulier A
,
Poiteau L
,
Rosa I
,
Hézode C
,
Roudot-Thoraval F
,
Pawlotsky JM
, et al.
Dried Blood Spots: A Tool to Ensure Broad Access to Hepatitis C Screening, Diagnosis, and Treatment Monitoring. J Infect Dis. 2016 Apr;213(7):1087–95. https://doi.org/10.1093/infdis/jiv423
33.
van Deursen P
,
Oosterlaken T
,
Andre P
,
Verhoeven A
,
Bertens L
,
Trabaud MA
, et al.
Measuring human immunodeficiency virus type 1 RNA loads in dried blood spot specimens using NucliSENS EasyQ HIV-1 v2.0. J Clin Virol. 2010 Feb;47(2):120–5. https://doi.org/10.1016/j.jcv.2009.11.021
34.
Abe K
,
Konomi N
. Hepatitis C virus RNA in dried serum spotted onto filter paper is stable at room temperature. J Clin Microbiol. 1998 Oct;36(10):3070–2. https://doi.org/10.1128/JCM.36.10.3070-3072.1998
35.
Catlett B
,
Carrera A
,
Starr M
,
Applegate TL
,
Lowe P
,
Grebely J
, et al.
Performance evaluation of the Hologic Aptima HCV Quant Dx assay for detection of HCV RNA from dried blood spots. J Clin Virol. 2019 Mar;112:40–4. https://doi.org/10.1016/j.jcv.2019.01.010
36.
Solmone M
,
Girardi E
,
Costa F
,
Pucillo L
,
Ippolito G
,
Capobianchi MR
. Simple and reliable method for detection and genotyping of hepatitis C virus RNA in dried blood spots stored at room temperature. J Clin Microbiol. 2002 Sep;40(9):3512–4. https://doi.org/10.1128/JCM.40.9.3512-3514.2002
37.
Moradpour D
,
Fehr J
,
Semela D
,
Rauch A
,
Müllhaupt B
. Treatment of Chronic Hepatitis C – January 2021 Update Expert Opinion SASL, SSG and SSI. Available from: https://www.sginf.ch/files/sasl-ssg-ssi_eos_hepc_jan2021_1.pdf (Accessed: 2021 May 20)
38.
European Association for the Study of the Liver Recommendations on treatment of hepatitis C: final update of the series. J Hepatol. 2020;73:1170–218. https://doi.org/10.1016/j.jhep.2020.08.018
39.
Ghany MG
,
Morgan TR
. AASLD-ISDA Hepatitis C Guidance Panel. Hepatitis C Guidance 2019 Update: AASLD-IDSA Recommendations for testing, managing, and treating hepatitis C virus infection. Hepatology. 2020;71:686–721. https://doi.org/10.1002/hep.31060
40.
Hajarizadeh B
,
Grady B
,
Page K
,
Kim AY
,
McGovern BH
,
Cox AL
, et al.; InC3 Study Group
. Patterns of hepatitis C virus RNA levels during acute infection: the InC3 study. PLoS One. 2015 Apr;10(4):e0122232. https://doi.org/10.1371/journal.pone.0122232
41.
Bertisch B
,
Brezzi M
,
Negro F
,
Müllhaupt B
,
Ottiger C
,
Künzler-Heule P
, et al.; Swiss Hepatitis C Cohort Study
. Very Low Hepatitis C Viral Loads in Treatment-naive Persons: Do They Compromise Hepatitis C Virus Antigen Testing? Clin Infect Dis. 2020 Feb;70(4):653–9.
42. WHO guidelines on hepatitis B and C testing. Geneva: World Health Organization; 2017
43. Offizielle Website der nationalen Substitutionsstatistik, 2016-2021. Available from: https://www.substitution.ch/de/jahrliche_statistik.html&year=2020&canton=ag (Accessed: 2021 May 20)
44.
Buchanan R
,
Cooper K
,
Grellier L
,
Khakoo SI
,
Parkes J
. The testing of people with any risk factor for hepatitis C in community pharmacies is cost-effective. J Viral Hepat. 2020 Jan;27(1):36–44. https://doi.org/10.1111/jvh.13207
45.
Buchanan R
,
Hassan-Hicks P
,
Noble K
,
Grellier L
,
Parkes J
,
Khakoo SI
. Integrating community pharmacy testing for hepatitis C with specialist care. Clin Pharm. 2016;8(8):243–7.
46.
Kugelmas M
,
Pedicone LD
,
Lio I
,
Simon S
,
Pietrandoni G
,
Hepatitis C
. Hepatitis C Point-of-Care Screening in Retail Pharmacies in the United States. Gastroenterol Hepatol (N Y). 2017 Feb;13(2):98–104.
47.
Radley A
,
Tait J
,
Dillon JF
. DOT-C: A cluster randomised feasibility trial evaluating directly observed anti-HCV therapy in a population receiving opioid substitute therapy from community pharmacy. Int J Drug Policy. 2017 Sep;47:126–36. https://doi.org/10.1016/j.drugpo.2017.05.042
48.
Radley A
,
de Bruin M
,
Inglis SK
,
Donnan PT
,
Hapca A
,
Barclay ST
, et al.
Clinical effectiveness of pharmacist-led versus conventionally delivered antiviral treatment for hepatitis C virus in patients receiving opioid substitution therapy: a pragmatic, cluster-randomised trial. Lancet Gastroenterol Hepatol. 2020 Sep;5(9):809–18. https://doi.org/10.1016/S2468-1253(20)30120-5
49.
Wade AJ
. Can community pharmacists treat hepatitis C virus? Lancet Gastroenterol Hepatol. 2020 Sep;5(9):790–1. https://doi.org/10.1016/S2468-1253(20)30184-9
50.
Bajis S
,
Applegate TL
,
Grebely J
,
Matthews GV
,
Dore GJ
. Novel Hepatitic C Virus (HCV) Diagnosis and Treatment Delivery Systems: Facilitating HCV Elimination by Thinking Outside the Clinic. J Infect Dis. 2020 Nov;222 Suppl 9:S758–72. https://doi.org/10.1093/infdis/jiaa366
51.
Bundesamt für Gesundheit BAG
. (03/2019) Hepatitis C bei Drogenkonsumierenden: Richtlinien mit settingspezifischen Factsheets. Available from: https://www.bag.admin.ch/dam/bag/de/dokumente/mt/infektionskrankheiten/hepatitis-c/richtlinien-hepatitis-c-drogen.pdf.download.pdf/richtlinien-hepatitis-c-drogen-de.pdf (Accessed: 2021 May 20)
52.
Smookler D
,
Vanderhoff A
,
Biondi MJ
,
Valencia J
,
Ryan P
,
Karkada J
,
Hong R
,
Sattar I
,
Mandel E
,
Gjevori M
,
Casey J
,
Fletcher D
,
Shah H
,
Hansen BE
,
Capraru C
,
Janssen HLA
,
Lazarus JV
,
Feld JJ
. Reducing Read Time of Point-of-Care Test Does Not Affect Detection of Hepatitis C Virus and Reduces Need for Reflex RNA. Clin Gastroenterol Hepatol 2020:S1542-3565(20)31068-5. doi: https://doi.org/10.1016/j.cgh.2020.07.058. Epub ahead of print. PMID: 32763480.
53.
Pallarés C
,
Carvalho-Gomes Â
,
Hontangas V
,
Conde I
,
Di Maira T
,
Aguilera V
, et al.
Performance of the OraQuick Hepatitis C virus antibody test in oral fluid and fingerstick blood before and after treatment-induced viral clearance. J Clin Virol. 2018 May;102:77–83. https://doi.org/10.1016/j.jcv.2018.02.016
54.
Khuroo MS
,
Khuroo NS
,
Khuroo MS
. Diagnostic accuracy of point-of-care tests for hepatitis C virus infection: a systematic review and meta-analysis. PLoS One. 2015 Mar;10(3):e0121450. https://doi.org/10.1371/journal.pone.0121450
55.
Chevaliez S
,
Poiteau L
,
Rosa I
,
Soulier A
,
Roudot-Thoraval F
,
Laperche S
, et al.
Prospective assessment of rapid diagnostic tests for the detection of antibodies to hepatitis C virus, a tool for improving access to care. Clin Microbiol Infect. 2016 May;22(5):459.e1–6. https://doi.org/10.1016/j.cmi.2016.01.009
56.
Lee SR
,
Kardos KW
,
Schiff E
,
Berne CA
,
Mounzer K
,
Banks AT
, et al.
Evaluation of a new, rapid test for detecting HCV infection, suitable for use with blood or oral fluid. J Virol Methods. 2011 Mar;172(1-2):27–31. https://doi.org/10.1016/j.jviromet.2010.12.009
57.
The Kirby Institute
. Monitoring hepatitis C treatment uptake in Australia (Issue 10). The Kirby Institute, UNSW Sydney, NSW, Australia, June 2019 (available online at: https://kirby.unsw.edu.au/report/monitoring-hepatitis-c-treatment-uptake-australia-issue-10-june-2019). For more information, contact Dr Behzad Hajari (bhajarizadeh@kirby.unsw.edu.au) or Professor Greg Dore (gdore@kirby.unsw.edu.au).
58.
Wade AJ
,
Doyle JS
,
Gane E
,
Stedman C
,
Draper B
,
Iser D
, et al.
Outcomes of Treatment for Hepatitis C in Primary Care, Compared to Hospital-based Care: A Randomized, Controlled Trial in People Who Inject Drugs. Clin Infect Dis. 2020 Apr;70(9):1900–6. https://doi.org/10.1093/cid/ciz546
59.
Palmer AY
,
Wade AJ
,
Draper B
,
Howell J
,
Doyle JS
,
Petrie D
, et al.
A cost-effectiveness analysis of primary versus hospital-based specialist care for direct acting antiviral hepatitis C treatment. Int J Drug Policy. 2020 Feb;76:102633. https://doi.org/10.1016/j.drugpo.2019.102633
60.
Loustaud-Ratti V
,
Debette-Gratien M
,
Carrier P
. European Association for the Study of the Liver and French hepatitis C recent guidelines: the paradigm shift. World J Hepatol. 2018 Oct;10(10):639–44. https://doi.org/10.4254/wjh.v10.i10.639
61.
Ryan P
,
Valencia J
,
Cuevas G
,
Troya J
,
Ramon C
,
Rodríguez A
, et al.
HCV screening based on dried blood samples and linkage to care in people who use drugs: A prospective study. Int J Drug Policy. 2021 Jun;92:103134. https://doi.org/10.1016/j.drugpo.2021.103134
62.
WHO Prequalification of In Vitro Diagnostics
. Public Report (June/2016, version 2.0), Product: Xpert® HIV-1 Qual Assay, WHO reference number: PQDx 0259-070-00, (available online at: https://www.who.int/diagnostics_laboratory/evaluations/pq-list/hiv-vrl/160613PQPublicReport_0259-0700-00_XpertQualHIV_v2.pdf)
Appendix: Supplementary figures
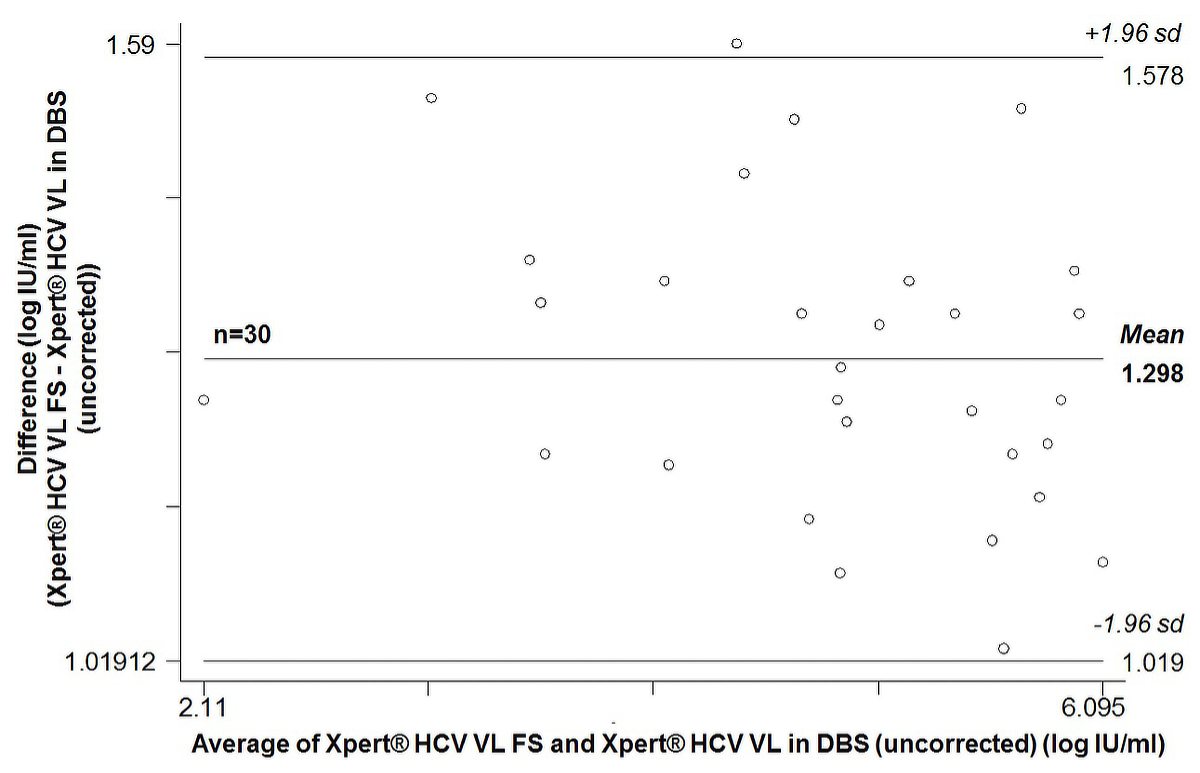
Figure S1 Bland-Altman plot of 30 paired quantifiable HCV RNA measurements: Xpert® HCV Viral Load Fingerstick (VL FS) test versus Xpert® HCV Viral Load (VL) test in dried blood spots produced with 100 µl capillary whole blood (uncorrected results).
sd = standard deviation; average bias: 1.298 (95% CI 1.246–1.350); 95% limits of agreement 1.019–1.578; Pitman’s test of difference in variance r = –0.283, p = 0.161
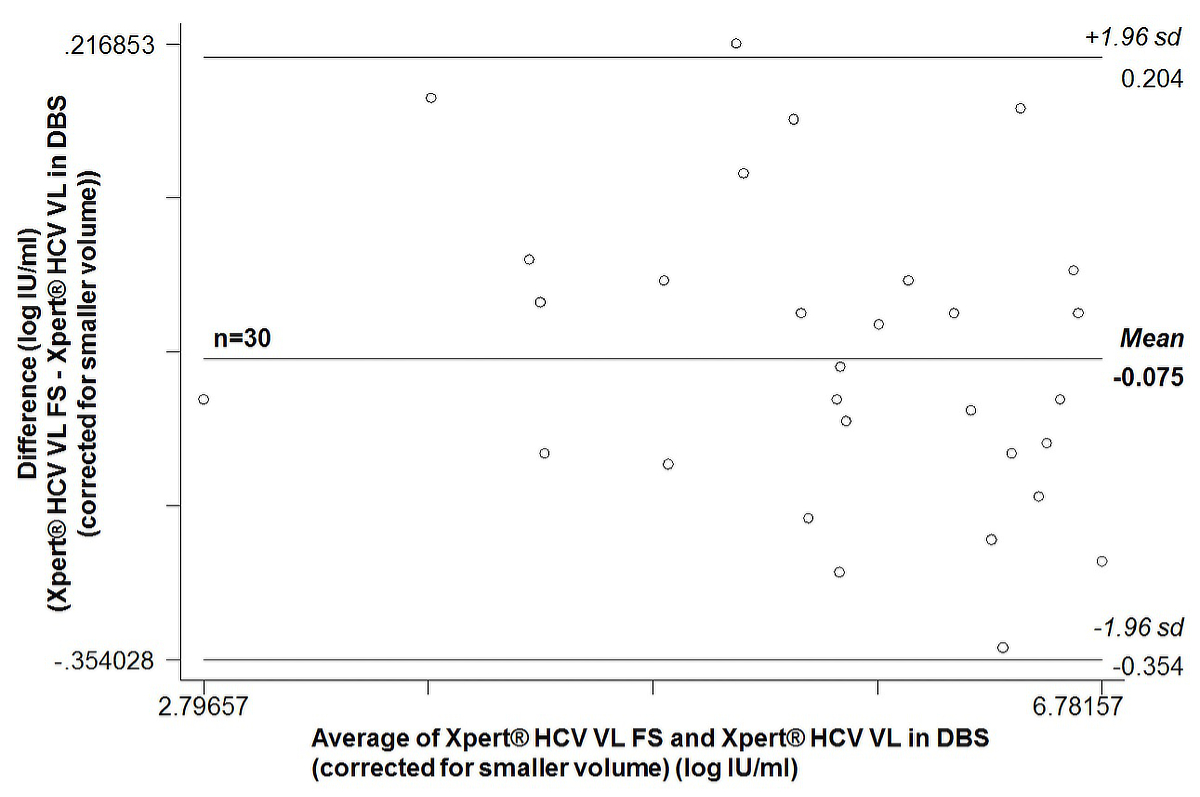
Figure S2 Bland-Altman plot of 30 paired quantifiable HCV RNA measurements: Xpert® HCV Viral Load Fingerstick (VL FS) test versus Xpert® HCV Viral Load (VL) test in dried blood spots produced with 100 µl capillary whole blood (results corrected for the smaller plasma volume).
sd = standard deviation; average bias: –0.075 (95% CI –0.127 to –0.023); 95% limits of agreement –0.354 to 0.204; Pitman’s test of difference in variance r = -0.283, p = 0.161




