PCR performance in the SARS-CoV-2 Omicron variant of concern?
DOI: https://doi.org/10.4414/SMW.2021.w30120
César M. J. A.
Metzgera, Reto
Lienhardbc, Helena M. B.
Seth-Smithde, Tim
Roloffde, Fanny
Wegnere, Jonas
Siebera, Michael
Belf, Gilbert
Greubcg, Adrian
Eglicde
aFederal Office for Civil Protection, Spiez Laboratory, Spiez, Switzerland
bADMed Microbiologie, La-Chaux-de-Fonds, Switzerland
cCommission of Clinical Microbiology (CCCM) of the Swiss Society of Microbiology
dClinical Bacteriology and Mycology, University Hospital Basel, Basel, Switzerland
eApplied Microbiology Research, Department Biomedicine, University of Basel, Basel, Switzerland
fFederal Office of Public Health, Bern, Switzerland
gInstitute for Medical Microbiology, University Hospital Lausanne, Lausanne, Switzerland
Dr. César Metzger
Federal Office for Civil Protection
Spiez Laboratory
CH-3700 Spiez
cesar.metzger[at]babs.admin.ch
and
Prof. Adrian Egli
Clinical Bacteriology and Mycology
University Hospital Basel
CH-4031 Basel
adrian.egli[at]usb.ch
Summary
The new SARS-CoV-2 Omicron variant (B.1.1.529) has been recently declared a Variant of Concern due to a series of important mutations in the viral spike protein and especially in the receptor-binding domain. While investigations into the spread of this new variant are ongoing, the first cases have been detected in Switzerland. Important questions have been raised: (1) Will the PCR assays commonly used to detect SARS-CoV-2 still work for the Omicron variant? (2) Can specific PCR features, e.g. S-gene dropout, be used to identify potential Omicron samples?
In this minireview we provide current knowledge on the Omicron variant and guidance on its PCR validation.
Introduction
Over the past 24 months, SARS-CoV-2 has continuously evolved into various defined viral lineages. Each lineage is characterized by a combination of specific nucleotide polymorphisms (SNPs) [1]. Some SNPs result in amino acid changes and may lead to functional adaptations that are beneficial for the virus in its interaction with the host, e.g. by increasing the affinity of the receptor-binding domain (RBD) of the viral spike protein (S protein) to the host entry point, the angiotensin-converting enzyme 2 (ACE2) receptor [2], resulting in a higher infectivity and a greater likelihood of successful transmission. A second important adaptation, also resulting in increased transmission, is immune evasion towards neutralizing antibodies [3, 4]. However, many polymorphisms remain silent and do not change the amino acid sequence (so-called synonymous mutations). Nevertheless, synonymous mutations may also have consequences for viral transmission: as rapid recognition of SARS-CoV-2 infection is a cornerstone of infection control measures, viral evolution in genomic regions used as diagnostic target sites may occur. This is often neglected, but is an important selective pressure for viruses [5]. To avoid PCR assay failure, highly conserved sites are often chosen as PCR targets, but these can be challenging to identify for novel pathogens. To offset this risk, multiple PCR targets can be used within a single assay to compensate for single target failures [6]. In this minireview, we discuss the consequences of viral evolution on the performance of commonly used SARS-CoV-2 diagnostic assays in the context of the newly discovered Omicron SARS-CoV-2 variant. We provide guidance on how to diagnose and process suspected cases of the Omicron variant.
Major steps in viral evolution
In summer 2020, a new variant, B.1.177 (Nextstrain 20E), emerged and spread rapidly in Europe [7]. This was followed by an even more efficient displacement by the Alpha variant (Pango Lineage B.1.1.7, Nextstrain 20I) in December 2020 to January 2021, driven by a higher binding affinity towards the ACE2 receptor [8]. The Alpha variant was rapidly declared a Variant of Concern (VOC). This lineage shows a series of important adaptations in the viral S protein, resulting in the previously mentioned increased binding affinity towards the host cell. From a diagnostic point of view, the del69/70 mutation was especially interesting as it resulted in S-gene dropout due to failure of the diagnostic target in some commercial PCR assays, such as the TaqPath assay by Thermo Fisher Scientific. This S-gene dropout could be used, during this period, to screen for potential Alpha variants prior to the development and implementation of further virus lineage-specific PCR assays and broad scale whole genome sequencing [9]. In Switzerland, the Federal Office of Public Health (FOPH) and the National Reference Centre for Emerging Viral Infections (CRIVE) coordinated the introduction of screening with lineage-specific PCR assays followed by whole genome sequencing for confirmation between December 2020 and March 2021. Backward contact tracing was used, with the intention of reducing and slowing down the spread of the Alpha variant [10]. In May 2021, the Delta variant, again with higher transmission rates, replaced Alpha within a few weeks [11] and has since been the dominant viral lineage around the globe [12].
The introduction of Omicron
On 26th November 2021, the World Health Organization (WHO) declared the emerging SARS-CoV-2 Omicron variant (Pango Lineage B.1.1.529 and Nextstrain Clade 21K) as another VOC [13]. The European Centre for Disease Prevention and Control (ECDC) rapidly published a threat assessment briefing [14]. This new member of the VOC family has an unusually high number of amino acid changes and deletions in the S protein, particularly in the RBD. Table 1 summarizes the key mutations found in the Omicron variant [17]. These mutations may potentially result in immune evasion, favourable transmission characteristics, and diagnostic evasion. The exact effects on immune evasion and favourable transmission are currently being evaluated. Probable transmission between two fully vaccinated persons has been reported in Hong Kong [15] and matches our own experience. New data from the Sigal lab suggest a 40-fold reduction in neutralizing capacity against the Omicron variant after the Pfizer vaccine [16].
|
Protein
|
ORF1a; %
|
ORF1b; %
|
S; %
|
E; %
|
M; %
|
N; %
|
| Position 1 |
K856R; 99% |
P314L; 100% |
A67V; 99% |
T9I, 97%
|
D3G, 97% |
P13L; NA
|
| Position 2 |
S2083I; 89% |
I1566V; 91% |
Del69/70; 89%* |
|
Q19E, 96%
|
ERS31del; NA
|
| Position 3 |
Del2084; 89% |
|
T95I; 98% |
|
A63T, 93%
|
R203K, 98%
|
| Position 4 |
A2710T; 100% |
|
G142D; 84%
|
|
|
G204R, 98% |
| Position 5 |
T3255I; 100%
|
|
Del143/145; 84% |
|
|
|
| Position 6 |
P3395H; 99%
|
|
N211I; 87% |
|
|
|
| Position 7 |
Del3674/3676; 88%
|
|
Del212/212; 87% |
|
|
|
| Position 8 |
I3758V; 100% |
|
G339D; 98%
|
|
|
|
| Position 9 |
P4715L; NA (B.1)
|
|
S371L; 98% |
|
|
|
| Position 10 |
I5967V; NA
|
|
S373P; 98%
|
|
|
|
| Position 11 |
|
|
S375F; 92%
|
|
|
|
| Position 12 |
|
|
K417N; NA
|
|
|
|
| Position 13 |
|
|
N440K; NA
|
|
|
|
| Position 14 |
|
|
S477N; 91%
|
|
|
|
| Position 15 |
|
|
T478K; 91%
|
|
|
|
| Position 16 |
|
|
E484A; 91%
|
|
|
|
| Position 17 |
|
|
Q493R; 94%
|
|
|
|
| Position 18 |
|
|
G496S; 94% |
|
|
|
| Position 19 |
|
|
Q498R; 94%
|
|
|
|
| Position 20 |
|
|
N501Y; 93%
|
|
|
|
| Position 21 |
|
|
Y505H; 91%
|
|
|
|
| Position 22 |
|
|
T547K; 100% |
|
|
|
| Position 23 |
|
|
D614G; 100% (B.1)
|
|
|
|
| Position 24 |
|
|
H655Y; 100%
|
|
|
|
| Position 25 |
|
|
N679K; 93%
|
|
|
|
| Position 26 |
|
|
P681H; 92%
|
|
|
|
| Position 27 |
|
|
N764K; NA
|
|
|
|
| Position 28 |
|
|
D796Y; 98% |
|
|
|
| Position 29 |
|
|
N856K; 98% |
|
|
|
| Position 30 |
|
|
Q954H; 98%
|
|
|
|
| Position 31 |
|
|
N969K; 97%
|
|
|
|
| Position 32 |
|
|
L981F; 95% |
|
|
|
The new Omicron variant was detected in 13 samples in the Basel region (since 30th October 2021). Notably, three sequence-confirmed cases in Basel occurred in fully vaccinated people via a fully vaccinated index patient. Omicron was also found in six samples in the Geneva region (since 1st December 2021), Switzerland. As of December 8th 2021, more than 15 sequences of the Omicron lineage have been detected in Switzerland (personal communication with FOPH) with community transmission and 595 sequences have been detected globally since its identification [17].
Knowledge on effects on PCR targets
Concern has been raised about the performance of commercial and in-house developed SARS-CoV-2 specific PCR assays with this new variant due to its high number of mutations [18]. In addition, the potential partial detection failure of some assays (that is, when a multiple target assay returns a positive result for only a few targets and a low or negative result for other targets) may be used to detect potential Omicron cases, as was the case with the Alpha variant.
In general, molecular diagnostic assays should target conserved sites (i.e. the genomic sequences least likely to accumulate mutations over time). At this stage in the SARS-CoV-2 pandemic, an unprecedented number of genomes are available and candidate conserved sites suitable for diagnostics can be readily identified. The chosen diagnostic targets should be reviewed on a regular basis by manufacturers and by laboratories to ensure continued efficient primer binding in currently circulating variants.
Unfortunately, companies rarely or never disclose detailed information on their assays’ diagnostic targets due to intellectual property concerns. Especially in an emerging situation such as the spread of a new VOC like Omicron, this has important consequences. The diagnostic laboratories do not have immediate information on the diagnostic performance of the commercial assays in use and critical cases early in the transmission chain may be missed. When companies rapidly make target information available, early action to ensure continued proper detection of VOCs can be taken. Incentives to promote such essential, rapid collaboration between manufacturers, laboratories and governmental bodies should be implemented in times of exceptional crisis such as the current pandemic. In cases where there is a critical need to understand the exact structure of the targets, a system of disclosure to a predetermined governmental body tasked with the in silico confirmation of diagnostic performance as an intermediary may be a promising avenue to consider, as this would ensure that companies’ intellectual property is protected.
We previously collected information on the PCR assays most commonly used by laboratories throughout Switzerland and Liechtenstein and determined which assays may be subject to target dropout or hindered performance with the Alpha variant [19]. In the context of the Omicron variant, we have re-examined previously obtained information and contacted the relevant manufacturers of the SARS-CoV-2 PCR assays available in Switzerland and Liechtenstein to obtain statements regarding PCR functionalities. Currently, the 39 most frequently used SARS-CoV-2 PCR tests in Switzerland and Liechtenstein target many genomic loci, including the ORF1ab region (N = 16), the RdRp gene (N = 13), the S gene (N = 8), the E gene (N = 11), the N gene (N = 32) and, in one case, the M gene. Figure 1 provides the PCR target analytes of SARS-CoV-2 specific assays commonly used in Switzerland and Liechtenstein and the currently available information concerning the impact of the Alpha and Omicron variants on the successful detection of the target analytes. Only two of the eight assays targeting the S gene appear to show S-gene dropout with the Omicron variant.
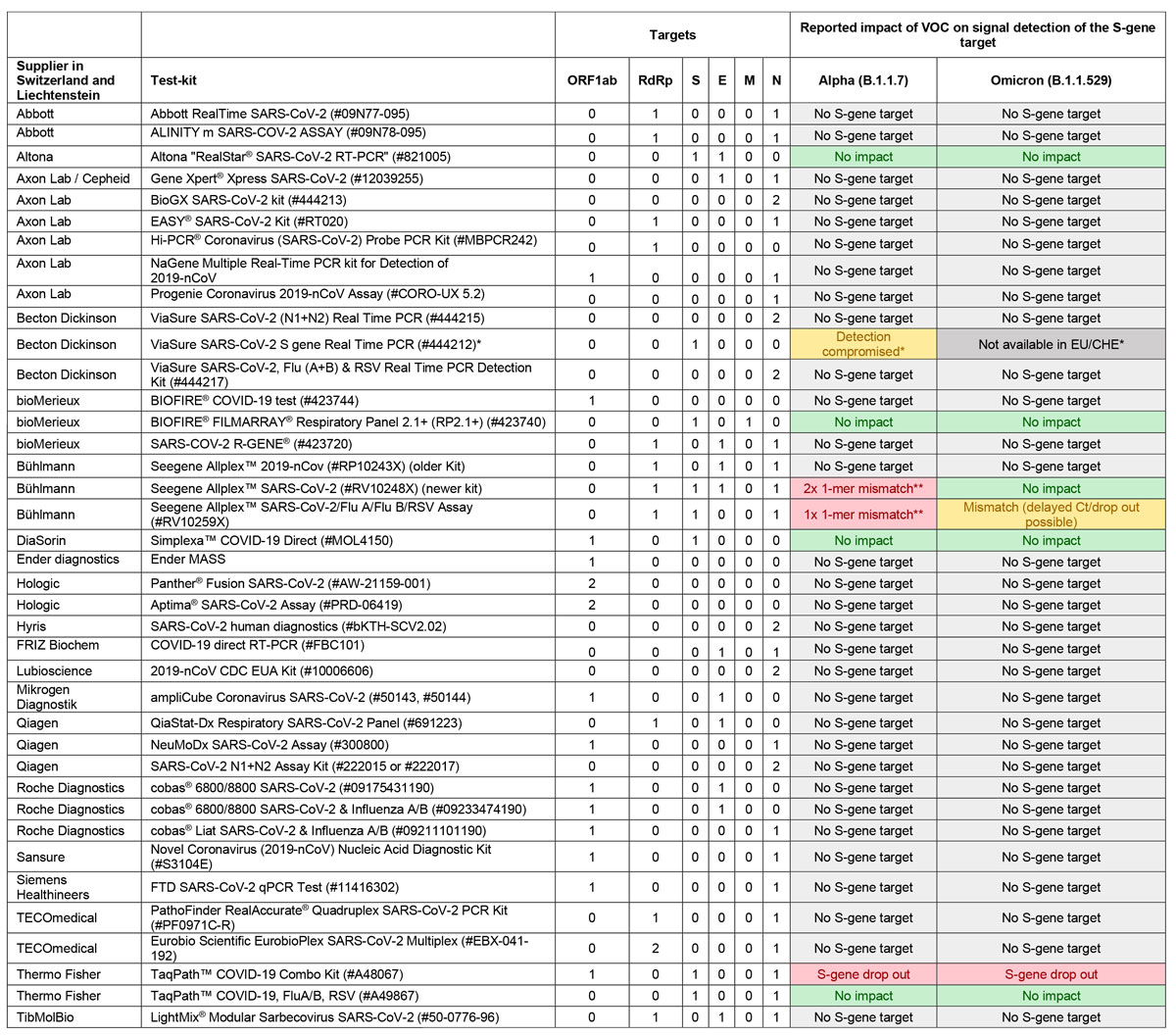
Figure 1 Selection of available PCR assays and the impacts on their performance of the Alpha and Omicron variants.
* PCR-Kit no longer available on the Swiss market.
** Mismatch not detectable because both the S gene and the RdRp gene targets are detected using the same fluorescence channel.
Given the many SNPs localized on the sequence encoding for the S protein, and concentrated on the RBD in particular, it is likely that some of the serology assays utilizing the wildtype S protein may exhibit decreased sensitivity. However, precise data are still lacking.
Surveillance and confirmation of Omicron
Due to the high demand for SARS-CoV-2 PCR testing throughout Switzerland and Liechtenstein caused by the ongoing epidemic surge with the currently dominating Delta variant, additional testing of all SARS-CoV-2 PCR positive cases seems unrealistic. Although systematic specific retesting would be ideal for surveillance of the spread of the new Omicron variant, the burden for most laboratories currently appears too high for this to happen. Although some laboratories already use assays with the potential for S-gene dropout (notably, this is how the first two Swiss cases, in the region of Basel, were detected), the majority of laboratories use different assays. With only a few laboratories using PCR assays which can indicate the Omicron variant through S-gene dropout, this would not generate a tight enough screening net. Notably, a new sub-lineage of Omicron, BA.2 (for more details see below), does not show S-gene dropout. Additional testing of targeted cases with epidemiological risk factors, such as travel history to high endemic areas or recent contact with an Omicron positive patient, could be implemented until the case load grows too large and community transmission is known to be widespread. This testing strategy would involve either a variant-specific PCR, in laboratories where this is already in use or has been accredited, or an initial diagnostic SARS-CoV-2 PCR without variant-specific features, followed by a variant-specific PCR. A commonly used target for the specific detection of the Alpha variant in the past was the N501Y-specific PCR. As Omicron shares the same N501Y mutation that allowed the detection of Alpha and the Alpha variant’s prevalence has become negligible, this is an option which should be considered (see figure 2). All such tests would need to be re-validated, as the number of mutations in this region of the Omicron genome may prevent primer binding in some assays or lead to reduced efficacy, preventing the detection of samples with low viral loads. Several alternative targets, such as L417N- or E484A-specific PCR assays, may also prove to be valuable options, as these targets are specific to Omicron. Recently, Erster and colleagues published a comparison of four RT-PCR assays specifically for the Omicron variant which target the deletion in the spike gene at positions 22194-22196 and the deletion in the N gene at positions 28362-28371 [20].
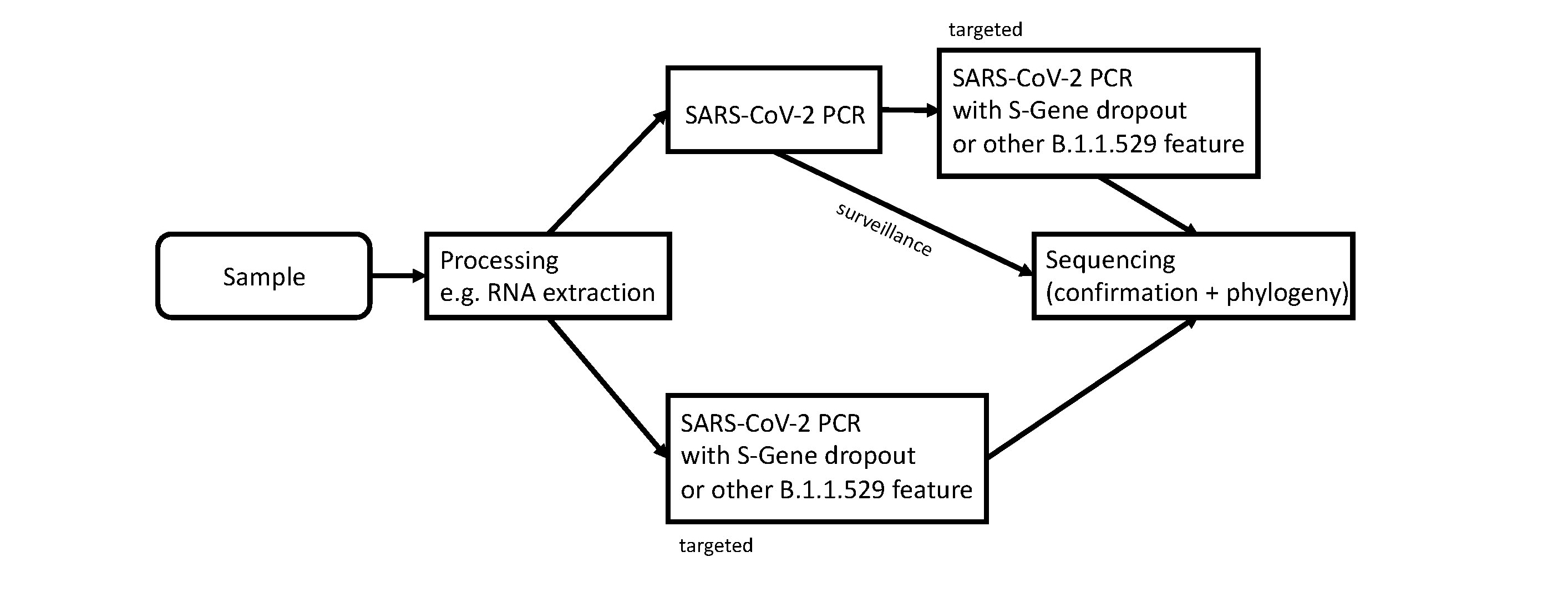
Figure 2 Workflow for screening and confirmation of the SARS-CoV-2 Omicron variant. “Surveillance” indicates the routine workflow for how Omicron variants could be detected by the sequencing surveillance program coordinated via CRIVE and the FOPH. “Targeted” indicates a suspected case of an Omicron variant which should be confirmed via sequencing.
For definitive confirmation of the Omicron variant, we recommend whole genome sequencing on one of the platforms by Oxford Nanopore Technology, Thermo Fisher Scientific or Illumina. Whole genome sequencing allows not only the confirmation of all mutations and clear lineage identification, but also provides valuable molecular epidemiological information. As an example, the first two cases recently detected in the region of Basel already show slight diversity. Whole genome sequencing therefore provides valuable epidemic intelligence information. Phylogenetic analysis of all the Omicron VOC sequences available in GISAID’s (www.gisaid.org) EpiCoV database indicates that similar ancestral sequences have been detected in several countries [14, 17]. Notably, this similarity suggests a low diversity of the Omicron variants in circulation and therefore a rapid spread.
Sanger sequencing of the S gene (and additional viral genes) may also be used to confirm the new variant. Sanger sequencing is more cost efficient than whole genome sequencing, but less efficient for generating molecular epidemiological connections as only parts of the genome are sequenced. Both Sanger and next generation sequencing approaches use PCR primers, which need to be tested for functional gaps, especially in heavily altered lineages such as the new Omicron variant.
Whole genome sequencing also relies on a set of primers. The ARTIC protocol is a commonly used set of primers which allows the viral genome to be sequenced. In silico analysis showed that amplicon 76 of the ARTIC v4 protocol could fail for the Omicron VOC due to primer-template mismatches. This can lead to missed calls at spike protein residues 417, 440 and 446. If this region is not covered, other characteristic mutations can be used to identify the variant [14]. Updated ARTIC v4.1 primers, designed to more effectively cover amplicon 76 and a few other regions of the Omicron variant, have just been released [21]. However, based on our experience, we conclude that the Artic v4 protocol allows sequencing and confirmation of the Omicron variant. We strongly recommend that sequencing data is immediately shared via the Swiss Pathogen Surveillance Platform (www.spsp.ch) and to public data repositories. This helps to track the viral evolution.
Based on published genomes, it has just been announced that Omicron (B.1.1.529) has been split into two sub-lineages: BA.1 and BA.2 at pangoLearn. It will typically take two to three days until all the lineage assignments have been run retrospectively and are propagated into databases such as outbreak.info (not available at this stage). Specific mutations for the sub-lineages BA.1 and BA.2 can be seen at Github (https://github.com/cov-lineages/pango-designation/issues/361). We highlighted in table 1 that the targets S:T478K, S:E484A and N501Y are conserved across BA.1 and BA.2. These targets have already been used for Alpha and might be easy to implement for diagnostic labs. The new sub-lineage BA.2 does not carry the S:69/70del and therefore cannot be detected with S-gene dropout only.
Conclusion
The new Omicron VOC currently poses a detection challenge for diagnostic laboratories. At the moment there is uncertainty in the diagnostic performance of the available PCR assays, as all information obtainable at this early stage is based on companies’ in silico evaluations. For many of the systems, hardly any real-world evidence is available yet. Laboratories should share RNA extracts from positive cases to rapidly assess the performance of the available systems. In addition, the political demands for broad surveillance of the spread of the Omicron variant seem unrealistic at this point, as laboratories are already under strain from being required to perform large amounts of PCR testing due to the ongoing and strong Delta wave (currently 9,000–11,000 new cases per day in Switzerland and Liechtenstein [22] as of 3rd December 2021). We propose a combination of a targeted approach and complementarily continuing with the national surveillance program of using whole genome sequencing for the emergence of variants. This program should be continued for the next 1–2 years to provide sufficient sequencing information for vaccine development as well as the adaptation of diagnostic assays. The recent discovery and rapid spread of the Omicron variant has clearly shown that we need to remain on our guard.
Acknowledgements
The authors would like to thank the companies and suppliers of PCR tests who provided additional information about their targets and the impact of the B.1.1.529 / Omicron variant on assays with their PCR kits.
1.
Hodcroft E
. CoVariants. 2021. https://covariants.org/variants/21K.Omicron (accessed 06.12.2021).
2.
Lan J
,
Ge J
,
Yu J
,
Shan S
,
Zhou H
,
Fan S
, et al.
Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 May;581(7807):215–20. https://doi.org/10.1038/s41586-020-2180-5
3.
Lazarevic I
,
Pravica V
,
Miljanovic D
,
Cupic M
. Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses. 2021 Jun;13(7):1192. https://doi.org/10.3390/v13071192
4.
Zhang J
,
Xiao T
,
Cai Y
,
Lavine CL
,
Peng H
,
Zhu H
, et al.
Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021 Oct;374(6573):eabl9463. https://doi.org/10.1126/science.abl9463
5.
Filkins L
,
SoRelle JA
,
Schoggins J
,
Park JY
. Laboratory Action Plan for Emerging SARS-CoV-2 Variants. Clin Chem. 2021 Apr;67(5):720–3. https://doi.org/10.1093/clinchem/hvab020
6.
Babb de Villiers C
,
Blackburn L
,
Cook S
, et al.
SARS-CoV-2 variants. In: Diagnostics FfIN, editor.; 2021. p. 1.
7.
Hodcroft EB
,
Zuber M
,
Nadeau S
,
Vaughan TG
,
Crawford KH
,
Althaus CL
, et al.; SeqCOVID-SPAIN consortium
. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021 Jul;595(7869):707–12. https://doi.org/10.1038/s41586-021-03677-y
8.
Liu Y
,
Liu J
,
Plante KS
,
Plante JA
,
Xie X
,
Zhang X
, et al.
The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2021 Nov. https://doi.org/10.1038/s41586-021-04245-0
9.
Washington NL
,
White S
,
Barrett KM
,
Cirulli ET
,
Bolze A
,
Lu JT
. S gene dropout patterns in SARS-CoV-2 tests suggest spread of the H69del/V70del mutation in the US. medRxiv 2020: 2020.12.24.20248814. https://doi.org/10.1101/2020.12.24.20248814
10.
Goncalves Cabecinhas AR
,
Roloff T
,
Stange M
,
Bertelli C
,
Huber M
,
Ramette A
, et al.
SARS-CoV-2 N501Y Introductions and Transmissions in Switzerland from Beginning of October 2020 to February 2021-Implementation of Swiss-Wide Diagnostic Screening and Whole Genome Sequencing. Microorganisms. 2021 Mar;9(4):677. https://doi.org/10.3390/microorganisms9040677
11.
Mlcochova P
,
Kemp SA
,
Dhar MS
,
Papa G
,
Meng B
,
Ferreira IA
, et al.; Indian SARS-CoV-2 Genomics Consortium (INSACOG); Genotype to Phenotype Japan (G2P-Japan) Consortium; CITIID-NIHR BioResource COVID-19 Collaboration
. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021 Nov;599(7883):114–9. https://doi.org/10.1038/s41586-021-03944-y
12.
Campbell F
,
Archer B
,
Laurenson-Schafer H
,
Jinnai Y
,
Konings F
,
Batra N
, et al.
Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021 Jun;26(24). https://doi.org/10.2807/1560-7917.ES.2021.26.24.2100509
13.
Organization WH
. Update on Omicron. 2021. https://www.who.int/news/item/28-11-2021-update-on-omicron (accessed 02.12.2021 2021).
14. Control ECfDPa. European Centre for Disease Prevention and Control. Implications of the further emergence and spread of the SARS CoV 2 B.1.1.529 variant of concern (Omicron) for the EU/EEA first update. 2021. https://www.ecdc.europa.eu/en/publications-data/covid-19-threat-assessment-spread-omicron-first-update (accessed 03.12.2021 2021).
15.
Gu H
,
Krishnan P
,
Ng DY
,
Chang LD
,
Liu GY
,
Cheng SS
, et al.
Probable Transmission of SARS-CoV-2 Omicron Variant in Quarantine Hotel, Hong Kong, China, November 2021. Emerg Infect Dis. 2021 Dec;28(2). https://doi.org/10.3201/eid2802.212422
16.
Cele S
,
Jackson L
,
Khan K
, et al.
SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. MedRxiv 2021; 267417v1. https://doi.org/10.1101/2021.12.08.21267417
17.
Mullen JL
,
Tsueng G
,
Abdel Latif A
, et al.
Outbreak.info. 2020. https://outbreak.info/ (accessed 03.12.2021 2021).
18.
Vogels CB
,
Breban MI
,
Ott IM
,
Alpert T
,
Petrone ME
,
Watkins AE
, et al.; Brazil-UK CADDE Genomic Network; Network for Genomic Surveillance in South Africa
. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021 May;19(5):e3001236. https://doi.org/10.1371/journal.pbio.3001236
19.
Lienhard R
,
Metzger CM
,
Sieber J
, et al.
What does the UK variant tell the clinical microbiologists? Pipette. 2021;(2):3.
20.
ERSTER O
. Beth Din A, Asraf H, et al. SPECIFIC DETECTION OF SARS-COV-2 B.1.1.529 (OMICRON) VARIANT BY FOUR RT-qPCR DIFFERENTIAL ASSAYS. medRxiv 2021: 2021.12.07.21267293.
21. SARS-CoV-2 V4.1 update for Omicron variant. 01.12.2021. https://community.artic.network/t/sars-cov-2-v4-1-update-for-omicron-variant/342 (accessed 05.12.2021 2021).
22.
Health FOoP
. COVID-19 Switzerland. 2021. https://www.covid19.admin.ch/en/epidemiologic/case (accessed 05.12.2021 2021).

