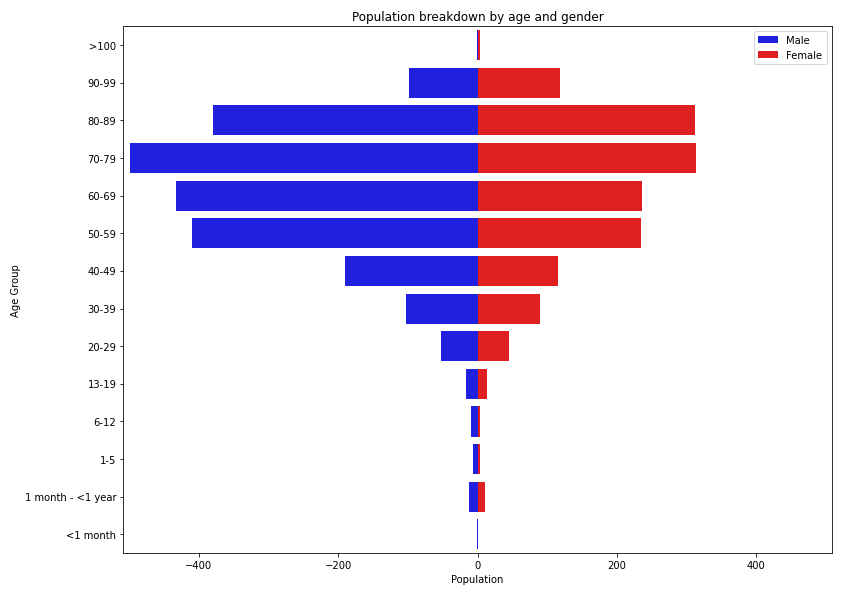
Figure 1 Population breakdown of the 3645 patients hospitalised with COVID 19 between 1 March and 31 August 2020, by age and gender.
DOI: https://doi.org/10.4414/smw.2021.20475
With almost 27 million confirmed cases, of which 70.5% recovered, and more than 850,000 deaths [1], the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) / coronavirus disease 2019 (COVID-19) pandemic has had a massive impact on all aspects of everyday life, including politics, travel and economics. The virus was initially identified in a cluster of Chinese adult patients with pneumonia of unknown cause [2], incriminating a large seafood and animal market in Wuhan City as the source of the outbreak; but rapidly spread via person-to-person transmission. Since then, 213 countries and territories have been affected [3].
On 30 January 2020, the World Health Organization (WHO) declared the outbreak a Public Health Emergency of International Concern (PHEIC). The WHO emphasised the urgent need to coordinate international efforts to investigate and better understand this novel coronavirus, to minimise the threat in affected countries and to reduce the risk of further international spread [4]. The WHO designed various study protocols for early investigation of the outbreak, including a global anonymised clinical data platform (the “nCoV Data Platform”) to enable state parties of the International Health Regulations (IHR) (2005) to share with the WHO anonymised clinical data and information related to patients with suspected or confirmed infections with SARS-CoV-2 (collectively “Anonymized nCoV Data”) [5].
The first patient with COVID-19 in Switzerland was reported on 25 February 2020 in the canton of Ticino, earlier than in most European countries due to its proximity to Italy and cross-border commuting. In order to prevent and monitor the spread of the disease in Switzerland, suspicion of COVID-19 cases by clinicians were reported to the Federal Office of Public Health (hereafter FOPH) through an obligatory declaration form [6]. The number of confirmed cases rose rapidly to more than 8000 positive cases and 66 deaths within only a month after the start of the epidemic in Switzerland. Compared with its neighbour countries, Switzerland has similarly suffered from the pandemic with a cumulative incidence of 496.5 confirmed cases per 100,000 habitants (Germany 264.7, France 449.9, Italy 448.4), but with an overall lower cumulative incidence of death at 200.9 deaths per 1,000,000 inhabitants (Germany 111.6, France 471.2, Italy 586.2) as of 4 September 2020 [7].
Early analysis and preparations by officials for the advent of the COVID-19 pandemic in Switzerland, including identification of risk groups and forecasting, were based on Chinese data. However, the Chinese population differs from the Swiss population, and such models were therefore misleading and inaccurate. Based on our experience with the hospital-based surveillance of influenza cases in Switzerland [8] and in collaboration with the FOPH, we developed a surveillance system of hospitalised COVID-19 cases in early February. We aimed to:
The aim of the current manuscript is to present a profile of patients hospitalised with COVID-19 during the first epidemic wave in Switzerland.
This study was submitted to and approved by the Geneva Ethics Committee (CCER) and by all hospitals’ local Ethics Committee through the Swissethics BASEC submission system, under reference 2020-00827.
The study was designed to capture most of the severe COVID-19 cases in Switzerland by including hospitalised patients from 20 large cantonal and university hospitals. To ease the process and to minimise the time taken to develop the system, we first contacted the participating hospitals of the influenza pilot surveillance system [8]; seven of which agreed to participate (Hôpitaux Universitaires de Genève, Kantonsspital St Gallen, Ospedaliero Cantonale Ticino, Hôpital du Valais, Centre Hospitalier Universitaire du Canton de Vaud, Universitätsspital Zürich, Universitäts-Kinderspital Zürich). Since patients were more likely to be sent to large hospitals with more capacity and better means to treat a novel disease, the system also captured patients from other large university and cantonal hospitals that agreed to participate: Universitätsspital Basel, Inselspital Bern, Kantonsspital Graubünden, Luzerner Kantonsspital (including Kinderspital Luzern), Spitäler Schaffhausen Kantonsspital, Thurgau Hospital Group, and smaller cantonal hospitals (Hôpital de Fribourg, Kantonsspital Aarau, Kantonsspital Winterthur, Kantonsspital Nidwalden, and Klinik Hirslanden Zürich). Three of the smaller hospitals (Fribourg, Aarau and Winterthur) contributed only paediatric cases. We also included three paediatric hospitals, separate from their adult counterparts: Universität Kinderspital Basel, Ostschweizer Kinderspital, and Kinderspital Zürich. The Kantonsspital St. Gallen also included cases from all public hospitals of the canton: they were included by the identical infection control team.
Patients, regardless of age or gender, with a polymerase chain-reaction (PCR) confirmed COVID-19 diagnosis and who were hospitalised for more than 24 hours were included in the database. The data were collected by infection control specialists or other physicians, dedicated study nurses or medical students of the participating centres under the supervision of physicians and stored anonymously in a secure REDCap database [9] located in the Clinical Research Centre of the University Hospitals of Geneva. Data entry started on 1 March 2020 and is still ongoing. Data were also collected retrospectively.
Data are collected on the case report form (CRF) developed for the influenza surveillance pilot [8], with relevant COVID-19 additions. This CRF is split in two parts: a compulsory part contains inclusion criteria, demographics, case declaration (classification, date of detection and date of symptoms, laboratory sample information), admission details (hospitalisation ward, origin of patient, severity at admission), and follow-up forms. The optional part describes the patient’s stay in more detail with descriptions of complications, intensive/intermediate care stays, treatment and risk factors in a clinical complementary information form. The CRF is presented in appendix 1.
Since the system was put in place when movement and contact tracing became difficult, we selected only part of the recommended WHO COVID-19 CRF [10] and of the FOPH obligatory declaration form [6]. We removed any information about travel to foreign countries or ethnicity. However, we kept employment in healthcare facilities as an important risk factor. The follow-up and daily forms of the WHO CRF were deemed too time consuming to complete by participating hospitals, and were therefore discarded in favour of the influenza CRF structure. Reporting of complications and underlying medical conditions were also similar to the influenza CRF with small additions of conditions shown to be of importance either as risk factors (e.g., hypertension, angiotensin converting-enzyme inhibitor treatment, obesity), or as consequences of the SARS-CoV-2 infection (acute respiratory distress syndrome (ARDS), thrombosis/embolism). To assess the severity of the disease at admission, we also added the CURB-65 score [11] for adults (CURB-65 = confusion, blood urea >42,8 mg/dl, respiratory rate> 30/min, blood pressure <90/60 mm Hg, age >65), and a similar severity score for children based on the expertise of the participating paediatric infectious disease specialists. Finally, we classified cases as community-acquired or nosocomial, by defining nosocomial cases to be those for which the onset of symptoms was more than 5 days after the patient’s admission date. This time limit was to account for the incubation time for the virus and is in line with the national recommendations of Swissnoso [12].
As a surveillance tool and because of the rapid growth of the epidemic, basic epidemiological information had to be reported within 48 hours, comprising at minimum the inclusion criteria, demographics and case declaration forms in order to report the new cases and their location with little delay. Additional information on the hospitalisation itself (admission, follow-up and clinical information) were recorded as soon as possible, depending on the severity of the epidemic and local issues.
The database records entries as COVID-19 episodes rather than per patient, with the possibility to link two episodes. A new episode was defined as readmission into hospital more than 30 days after the previous discharge. If a transfer between hospitals occurred, a new record was created in the destination hospital. The database and CRF implementation were tested by the participating sites at the time of deployment and tweaked on the basis of their feedback. Small additional changes were made as the database was already in production when new information about the disease became available (e.g., thrombosis/embolism).
The data presented in this manuscript were extracted early on 1 September 2020, to ensure all patients up to and including 31 August 2020 were presented.
The data were processed through a set of Python (version 3.8.4) scripts, which assessed their quality and removed 115 ineligible records. Patient level data (year of birth and gender) were taken as is and age was derived by using the date of inclusion in the system. Episode data were aggregated by summing the hospitalisations under a same record ID for questions “Yes/No/Unknown”, “Unknown” answers being treated as missing values, “Yes” as 1 and “No” as 0. The outcome of the episode was taken in the last recorded hospitalisation for that episode.
Finally, the week used in the figures of this manuscript is the week of interest for the system, defined as:
As of 31 August 2020, 3650 episodes in 3645 patients have been recorded in the database, with 66 patients hospitalised more than once for the same episode. Figure 1 shows the age and gender distribution of patients in the database and table 1 shows their baseline characteristics. Overall, 2168 (59.5%) patients were male and 1477 (40.5%) female, and the median age was 68 years (interquartile range [IQR] 54–79). Most were aged between 50 and 89 years (2778, 76.2%), followed by the age groups 40 to 49 years (299, 8.2%), 90 to 99 years (215, 5.9%) and 30 to 39 years (185, 5.1%). Among children, patients between 13 and 19 years and patients between a month and less than a year were predominant, with 24 (0.7%; 35.3% of all children) and 23 (0.6%; 33.8% of all children) patients, respectively.

Figure 1 Population breakdown of the 3645 patients hospitalised with COVID 19 between 1 March and 31 August 2020, by age and gender.
Table 1 Baseline characteristics of patients registered in the database.
|
Patients
(N = 3645) |
|||
|---|---|---|---|
| n | % | ||
| Gender |
Male | 2168 | 59.5 |
| Female | 1477 | 40.5 | |
| Age group |
<1 month | 2 | 0.1 |
| 1 month – <1 year | 23 | 0.6 | |
| 1–5 | 10 | 0.3 | |
| 6–12 | 9 | 0.2 | |
| 13–19 | 24 | 0.7 | |
| 20–29 | 95 | 2.6 | |
| 30–39 | 185 | 5.1 | |
| 40–49 | 299 | 8.2 | |
| 50–59 | 629 | 17.3 | |
| 60–69 | 663 | 18.2 | |
| 70–79 | 804 | 22.1 | |
| 80–89 | 682 | 18.7 | |
| 90–99 | 215 | 5.9 | |
| >100 | 5 | 0.1 | |
The majority of episodes were community acquired (3249, 89.0%), and 349 (9.6%) were nosocomial or of unknown source (52, 1.4%). Nosocomial cases were found less often in children (4.4% of all episodes in children versus 9.7% of all episodes in adults). Figure 2 shows the evolution of the number of episodes throughout the epidemic. The first episodes were recorded in week 2020-09, followed by a rapid rise to about 900 episodes in week 2020-13 before decreasing and stagnating from week 2020-20 onwards with few episodes declared. A small increase in week 2020-25 was observed. At the time of writing, the number of new cases had stabilised to around 30 per week.
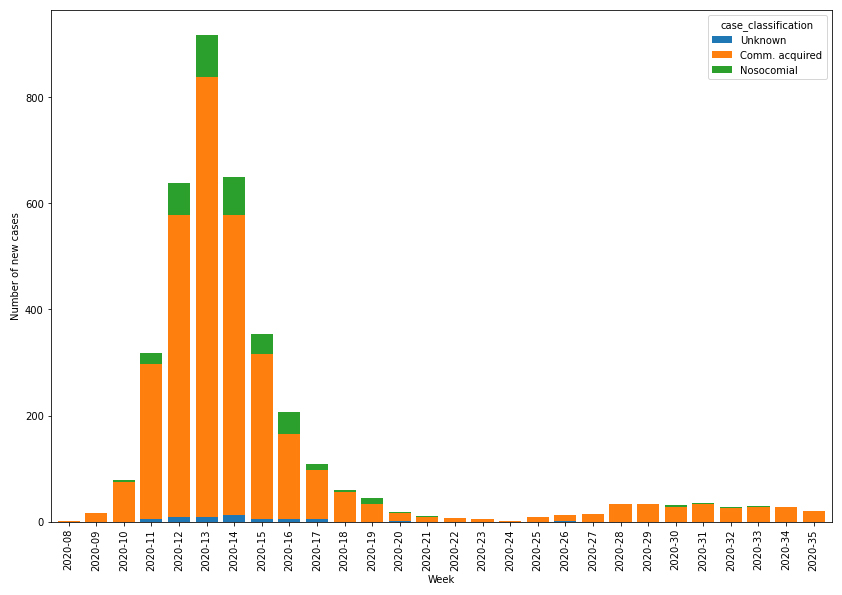
Figure 2 Distribution of SARS-CoV-2/COVID-19 episodes registered in the database by origin of infection.
The risk factors recorded, categorised into comorbidities (typically, chronic diseases), pregnancy status for women and history of smoking are split by age group for adults and children in tables 2 and 3 , respectively. The Charlson Comorbidity Index [13] was used to calculate an overall risk score, of which the median value was 3.0 (IQR 1–6) for both the overall population and adult patients. This score had a median value of 0.5 (IQR 0–1.75) among children.
Table 2 Characteristics of SARS-CoV-2/COVID-19 episodes, outcomes and related consequences (complications) encountered in registered episodes for adults (age groups 20–29 to >100).
|
Episodes
(N = 3582) |
|||
|---|---|---|---|
| n | % | ||
| Case classification |
Unknown | 47 | 1.3 |
| Community-acquired | 3189 | 89 | |
| Nosocomial | 346 | 9.7 | |
| Smoked in the last 10 years |
Unknown | 1537 | 42.9 |
| No | 1808 | 50.5 | |
| Yes | 237 | 6.6 | |
| Pregnancy (females aged between 15 and 50 only) |
Unknown | 381 | 65.8 |
| No | 163 | 28.2 | |
| Yes | 35 | 6 | |
| Had at least one comorbidity |
Unknown | 574 | 16 |
| Yes | 2401 | 67 | |
| No | 607 | 16.9 | |
| – Respiratory diseases |
Unknown | 3 | 0.1 |
| No | 1883 | 78.4 | |
| Yes | 515 | 21.4 | |
| – Asthma |
Unknown | 4 | 0.2 |
| No | 2193 | 91.3 | |
| Yes | 204 | 8.5 | |
| – Diabetes |
Unknown | 1 | 0 |
| No | 1740 | 72.5 | |
| Yes | 660 | 27.5 | |
| – Hypertension |
Unknown | 3 | 0.1 |
| No | 917 | 38.2 | |
| Yes | 1481 | 61.7 | |
| – Cardiovascular diseases |
Unknown | 4 | 0.2 |
| No | 1449 | 60.3 | |
| Yes | 948 | 39.5 | |
| – Renal diseases |
Unknown | 4 | 0.2 |
| No | 1897 | 79 | |
| Yes | 500 | 20.8 | |
| – Liver diseases |
Unknown | 3 | 0.1 |
| No | 2254 | 93.9 | |
| Yes | 144 | 6 | |
| – Neurological impairment |
Unknown | 8 | 0.3 |
| No | 2007 | 83.6 | |
| Yes | 386 | 16.1 | |
| – Haematological diseases with immunosuppression |
Unknown | 2 | 0.1 |
| No | 2342 | 97.5 | |
| Yes | 57 | 2.4 | |
| – Oncological diseases |
Unknown | 7 | 0.3 |
| No | 2079 | 86.6 | |
| Yes | 315 | 13.1 | |
| – Rheumatological diseases with immunosuppression |
Unknown | 2 | 0.1 |
| No | 2321 | 96.7 | |
| Yes | 78 | 3.2 | |
| – Dementia |
Unknown | 12 | 0.5 |
| No | 2191 | 91.3 | |
| Yes | 198 | 8.2 | |
| – Transplantation of solid organs |
Unknown | 4 | 0.2 |
| No | 2374 | 98.9 | |
| Yes | 23 | 1 | |
| – HIV positive |
Unknown | 45 | 1.9 |
| No | 2341 | 97.5 | |
| Yes | 15 | 0.6 | |
| – Under immunosuppressive treatment |
Unknown | 4 | 0.2 |
| No | 2284 | 95.1 | |
| Yes | 113 | 4.7 | |
| – Tuberculosis |
Unknown | 16 | 0.7 |
| No | 2363 | 98.4 | |
| Yes | 22 | 0.9 | |
| – Other diseases |
Unknown | 3 | 0.1 |
| No | 968 | 40.3 | |
| Yes | 1430 | 59.6 | |
| Had at least one complication |
Unknown | 587 | 16.4 |
| No | 345 | 9.6 | |
| Yes | 2650 | 74 | |
| – Ear/nose/throat (ENT) complications |
Unknown | 18 | 0.7 |
| No | 2439 | 92 | |
| Yes | 193 | 7.3 | |
| – Acute otitis media (included in ENT complications) |
Unknown | 3 | 1.6 |
| No | 182 | 94.3 | |
| Yes | 8 | 4.1 | |
| – Respiratory complications |
Unknown | 6 | 0.2 |
| No | 174 | 6.6 | |
| Yes | 2470 | 93.2 | |
| – Acute respiratory distress syndrome (included in respiratory complications) |
Unknown | 10 | 0.4 |
| No | 1903 | 77 | |
| Yes | 557 | 22.5 | |
| – Pneumonia (included in respiratory complications) |
Unknown | 19 | 0.8 |
| No | 289 | 11.7 | |
| Yes | 2162 | 87.5 | |
| – Pneumonia linked to COVID-19 (included with pneumonia) |
Unknown | 9 | 0.5 |
| No | 57 | 2.6 | |
| Yes | 2096 | 96.9 | |
| – Cardiovascular complications |
Unknown | 32 | 1.2 |
| No | 1987 | 75 | |
| Yes | 631 | 23.8 | |
| – Digestive complications |
Unknown | 41 | 1.5 |
| No | 2111 | 79.7 | |
| Yes | 498 | 18.8 | |
| – Liver complications |
Unknown | 28 | 1.1 |
| No | 2112 | 79.7 | |
| Yes | 510 | 19.2 | |
| – Renal complications |
Unknown | 28 | 1.1 |
| No | 1941 | 73.2 | |
| Yes | 681 | 25.7 | |
| – Neurological impairment complications |
Unknown | 44 | 1.7 |
| No | 2170 | 81.9 | |
| Yes | 436 | 16.5 | |
| – Osteo-articular complications |
Unknown | 34 | 1.3 |
| No | 2489 | 93.9 | |
| Yes | 127 | 4.8 | |
| – Thrombosis/embolism |
Unknown | 34 | 1.3 |
| No | 2406 | 90.8 | |
| Yes | 210 | 7.9 | |
| – Other bacterial infection |
Unknown | 38 | 1.4 |
| No | 2283 | 86.2 | |
| Yes | 329 | 12.4 | |
| – Other non-bacterial infection |
Unknown | 33 | 1.2 |
| No | 2485 | 93.8 | |
| Yes | 132 | 5 | |
| – Other complications |
Unknown | 19 | 0.7 |
| No | 1475 | 55.7 | |
| Yes | 1156 | 43.6 | |
| Stayed in ICU at least once |
Unknown | 604 | 16.9 |
| No | 2406 | 67.2 | |
| Yes | 572 | 16 | |
| Was treated against the COVID-19 infection |
Unknown | 611 | 17.1 |
| No | 1676 | 46.8 | |
| Yes | 1295 | 36.2 | |
| – Hydroxychloroquine |
No | 310 | 23.9 |
| Yes | 985 | 76.1 | |
| – Interferon |
No | 1293 | 99.8 |
| Yes | 2 | 0.2 | |
| – Lopinavir/ritonavir |
No | 620 | 47.9 |
| Yes | 675 | 52.1 | |
| – Remdesivir |
No | 1150 | 88.8 |
| Yes | 145 | 11.2 | |
| – Tenofovir |
No | 1294 | 99.9 |
| Yes | 1 | 0.1 | |
| – Ribavirin |
No | 1295 | 100 |
| Yes | 0 | 0 | |
| – Other treatment |
No | 1126 | 86.9 |
| Yes | 169 | 13.1 | |
| Outcome |
Deceased | 527 | 14.7 |
| Discharged and alive | 2923 | 81.6 | |
| Still hospitalised | 132 | 3.7 | |
HIV = human immunodeficiency virus; ICU = intensive care unit
Table 3 Characteristics of SARS-CoV-2/COVID-19 episodes, outcomes and related consequences (complications) encountered in registered episodes for children and teenagers (age groups <1 month to 13–19 years).
|
Episodes
(N = 68) |
|||
|---|---|---|---|
| n | % | ||
| Case classification |
Unknown | 5 | 7.4 |
| Community-acquired | 60 | 88.2 | |
| Nosocomial | 3 | 4.4 | |
| Smoked in the last 10 years |
Unknown | 44 | 64.7 |
| No | 22 | 32.4 | |
| Yes | 2 | 2.9 | |
| Pregnancy (female aged between 15 and 19 only) | Unknown | 11 | 64.7 |
| No | 6 | 35.3 | |
| Yes | 0 | 0 | |
| Had at least one comorbidity |
Unknown | 7 | 10.3 |
| Yes | 19 | 27.9 | |
| No | 42 | 61.8 | |
| – Respiratory diseases |
No | 15 | 78.9 |
| Yes | 4 | 21.1 | |
| – Asthma |
No | 15 | 78.9 |
| Yes | 4 | 21.1 | |
| – Diabetes |
No | 18 | 94.7 |
| Yes | 1 | 5.3 | |
| – Hypertension |
No | 18 | 94.7 |
| Yes | 1 | 5.3 | |
| – Cardiovascular diseases |
No | 18 | 94.7 |
| Yes | 1 | 5.3 | |
| – Renal diseases |
No | 17 | 89.5 |
| Yes | 2 | 10.5 | |
| – Liver diseases |
No | 17 | 89.5 |
| Yes | 2 | 10.5 | |
| – Neurological impairment |
No | 17 | 89.5 |
| Yes | 2 | 10.5 | |
| – Haematological diseases with immunosuppression |
No | 16 | 84.2 |
| Yes | 3 | 15.8 | |
| – Oncological diseases |
No | 16 | 84.2 |
| Yes | 3 | 15.8 | |
| – Rheumatological diseases with immunosuppression |
No | 19 | 100 |
| Yes | 0 | 0 | |
| – Dementia |
No | 19 | 100 |
| Yes | 0 | 0 | |
| – Transplantation of solid organs |
No | 19 | 100 |
| Yes | 0 | 0 | |
| – HIV positive |
No | 19 | 100 |
| Yes | 0 | 0 | |
| – Under immunosuppressive treatment |
No | 17 | 89.5 |
| Yes | 2 | 10.5 | |
| – Tuberculosis |
No | 19 | 100 |
| Yes | 0 | 0 | |
| – Other diseases |
No | 7 | 36.8 |
| Yes | 12 | 63.2 | |
| Had at least one complication |
Unknown | 9 | 13.2 |
| No | 30 | 44.1 | |
| Yes | 29 | 42.6 | |
| – Ear/nose/throat (ENT) complications |
No | 24 | 82.8 |
| Yes | 5 | 17.2 | |
| – Acute otitis media (included in ENT complications) |
No | 4 | 80 |
| Yes | 1 | 20 | |
| – Respiratory complications |
No | 13 | 44.8 |
| Yes | 16 | 55.2 | |
| – Acute respiratory distress syndrome (included in respiratory complications) |
Unknown | 1 | 6.2 |
| No | 11 | 68.8 | |
| Yes | 4 | 25 | |
| – Pneumonia (included in respiratory complications) |
Unknown | 1 | 6.2 |
| No | 4 | 25 | |
| Yes | 11 | 68.8 | |
| – Pneumonia linked to COVID-19 (included with pneumonia) |
Unknown | 1 | 9.1 |
| No | 0 | 0 | |
| Yes | 10 | 90.9 | |
| – Cardiovascular complications |
No | 23 | 79.3 |
| Yes | 6 | 20.7 | |
| – Digestive complications |
No | 22 | 75.9 |
| Yes | 7 | 24.1 | |
| – Liver complications |
No | 22 | 75.9 |
| Yes | 7 | 24.1 | |
| – Renal complications |
No | 26 | 89.7 |
| Yes | 3 | 10.3 | |
| – Neurological impairment complications |
No | 25 | 86.2 |
| Yes | 4 | 13.8 | |
| – Osteo-articular complications |
No | 29 | 100 |
| Yes | 0 | 0 | |
| – Thrombosis/embolism |
No | 26 | 89.7 |
| Yes | 3 | 10.3 | |
| – Other bacterial infection |
No | 29 | 100 |
| Yes | 0 | 0 | |
| – Other non-bacterial infection |
No | 27 | 93.1 |
| Yes | 2 | 6.9 | |
| – Other complications |
No | 15 | 51.7 |
| Yes | 14 | 48.3 | |
| Stayed in ICU at least once |
Unknown | 8 | 11.8 |
| No | 54 | 79.4 | |
| Yes | 6 | 8.8 | |
| Was treated against the COVID-19 infection |
Unknown | 7 | 10.3 |
| No | 57 | 83.8 | |
| Yes | 4 | 5.9 | |
| – Hydroxychloroquine |
No | 2 | 50 |
| Yes | 2 | 50 | |
| – Interferon |
No | 4 | 100 |
| Yes | 0 | 0 | |
| – Lopinavir/ritonavir |
No | 4 | 100 |
| Yes | 0 | 0 | |
| – Remdesivir |
No | 3 | 75 |
| Yes | 1 | 25 | |
| – Tenofovir |
No | 4 | 100 |
| Yes | 0 | 0 | |
| – Ribavirin |
No | 4 | 100 |
| Yes | 0 | 0 | |
| – Other treatment |
No | 1 | 25 |
| Yes | 3 | 75 | |
| Outcome |
Deceased | 0 | 0 |
| Discharged and alive | 66 | 97.1 | |
| Still hospitalised | 2 | 2.9 | |
HIV = human immunodeficiency virus; ICU = intensive care unit
In adults, comorbidities were recorded for 2401 (67.0%) episodes, with 1884 having more than one comorbidity. Among those, hypertension was the most frequent comorbidity, with 1481 (61.7%) occurrences, followed by chronic cardiovascular diseases (948, 39.5%) and diabetes (660, 27.5%). Chronic respiratory diseases were reported in 515 (21.4%) episodes, renal disease in 500 (20.8%) and neurological impairment in 386 (16.1%). Oncological pathologies (315, 13.1%), asthma (204, 8.5%), dementia (198, 8.2%), liver diseases (144, 6.0%) and immunosuppression (128, 5.3%) were registered less often, and other comorbidities were observed only sporadically (<5%). Information about comorbidities was missing for 574 (16.0%) episodes among adults.
In children, comorbidities were recorded for 19 (27.9%) episodes, with 9 having more than one comorbidity. Respiratory diseases and asthma were the most frequent comorbidities, occuring in 4 (21.1%) episodes each, followed by haematological and oncological pathologies (3, 15.8%), renal and liver diseases alongside neurological impairment and immunosuppression (2, 10.5%). Finally, diabetes, hypertension and cardiovascular diseases each occured in one (5.3%) episode. Information about comorbidities was missing for 7 (10.3%) episodes in children.
Tables 2 and 3 also show lists of complications encountered in COVID-19 episodes in adults and children, respectively. Out of the 3650 episodes, at least one complication due to the COVID-19 infection was reported in 2679 (73.4%), 2650 being in adults and 29 in children. Multiple complications occurred in 1961 adult episodes and 14 child episodes. Overall, 565 episodes with complications involved a stay on the intensive care unit (ICU), 560 of which were in adults.
In adults, respiratory complications were dominant, with 2470 (93.2%) occurrences. Pneumonia was found in 2162 episodes (87.5% of respiratory complications), of which 2096 (96.9%) were found to be caused solely by COVID-19. Acute respiratory distress syndrome (ARDS) was reported in 557 (22.5%) cases of respiratory complications. Renal complications were observed in 681 (25.7%) adult episodes, followed by cardiac (631, 23.8%), liver (510, 19.2%), digestive (498, 18.8%) and neurological (436, 16.5%) complications, and bacterial infections other than pneumonia (329, 12.4%). Other complications were present in less than 10% of the episodes and included thrombosis and embolisms (210, 7.9%), ear/nose/throat complications (ENT; 193, 7.3%; among which 8 [4.1%] were acute otitis media), non-bacterial infections (132, 5.0%) and osteo-articular complications (127, 4.8%).
In children, respiratory complications were also predominant with 16 (55.2%) occurrences. Pneumonia (11, 68.8%) was directly caused by COVID-19 in 10 (90.9%) cases of pneumonia. These comprised ARDS in 4 (25.0%) episodes in children. In contrast to adults, digestive and liver complications were the second most frequent complications, were reported in 7 (24.1%) children’s episodes with complications, followed by cardiac (6, 20.7%), ENT (5, 17.2%; of which one was a case of acute otitis media) and neurological (4, 13.8%) complications. Finally, renal complications and thrombosis/embolism were each reported in three episodes (10.3% of episodes with complications), and non-bacterial infections in 2 (6.9%). Other bacterial infections as well as osteo-articular complications were not reported in children.
Other types of complications were registered in 1170 episodes (43.6% of episodes with complications). Ageusia, anaemia and deconditioning syndromes were most often reported. Anosmia was reported in 4 (0.2%) COVID-19 episodes. The information was missing for 596 (16.3%) of the 3650 episodes.
The distribution of drugs prescribed against the COVID-19 infection can be found in tables 2 and 3 . In 1299 episodes (35.6% overall, 36.2% of episodes in adults and 5.9% of all episodes in children), one or often a combination of therapeutic agents were used. Hydroxychloroquine was the most prescribed treatment against COVID-19 (989, 76.1%; 985 in adults and 2 in children) followed by lopinavir/ritonavir (675, 52.0%; exclusively in adults). Remdesivir was the third most predominant drug used to treat COVID-19 (146, 11.2%; only once in children). Other treatments were prescribed rarely (interferon twice and tenofovir once), or not at all (ribavirin). Note that hydroxychloroquine was almost abandoned once no more recommended by international and national authorities.
Figure 3 shows the cumulative number of episodes, discharges and deaths reported in the database. Among the 3650 episodes, 2989 (81.9%) were reported be followed by discharge from the hospital, among which 2275 were discharged to their domicile, 230 to another hospital. 222 to a long-term care facility and 249 to other locations such as rehabilitation centres.
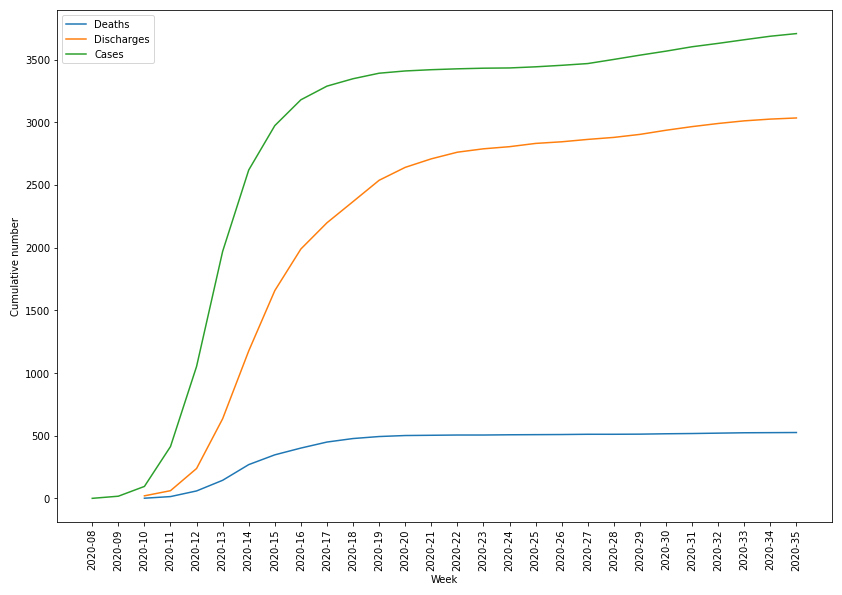
Figure 3 Cumulative number of episodes, discharges and deaths related to COVID-19 infection in the database.
Death was reported in 527 episodes (14.4%), none in children. The first death occurred in the week following the first recorded episode (2020-10). The number of reported deaths increased rapidly during the first 10 weeks, with only a small number of deaths occurring after week 2020-20. Figure 4 shows the number of death versus the time between the date of interest for the surveillance and the death date. Most deaths occurred in patients with comorbidities (431, 81.8%) within the first two weeks following their diagnosis, with a median time between diagnosis and death of 8 days (IQR 5–14 days). Overall, deaths were mostly in patients aged between 80 and 89 years (224, 42.5%) and in patients between 70 and 79 years (147, 27.8%) as seen in figure 5. No deaths were reported for patients below 30.
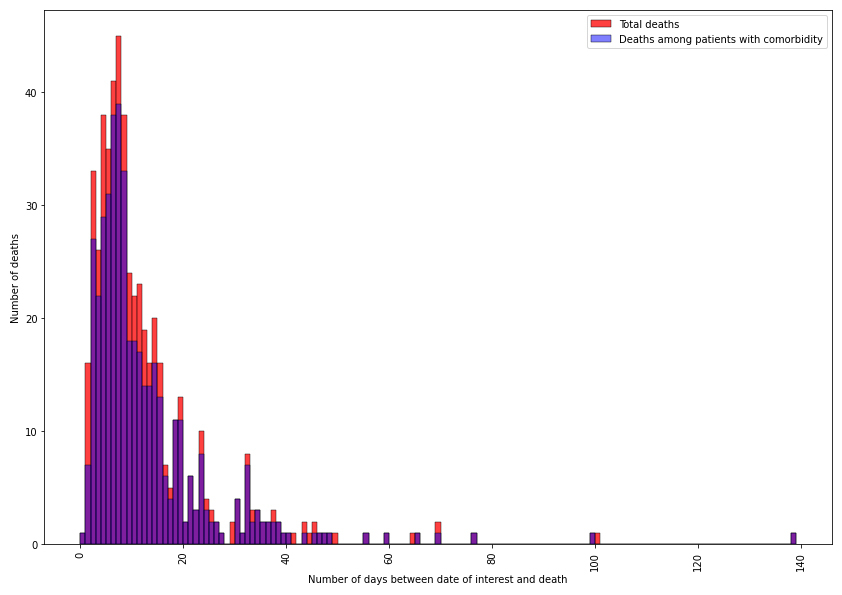
Figure 4 Distribution of deceased patients within the database.
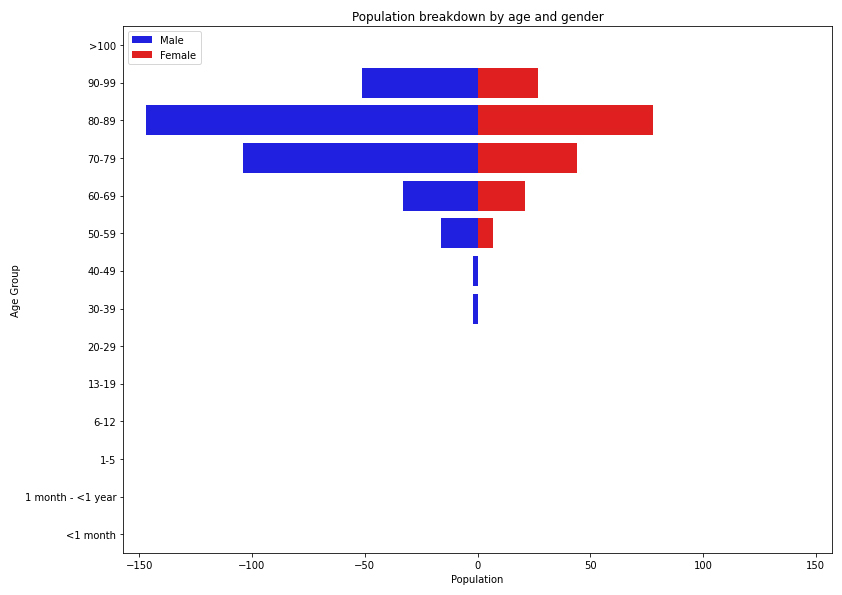
Figure 5 Population breakdown of the 527 deceased patients hospitalised with COVID 19 between 1 March and 31 August 2020 by age and gender.
Pregnancy was present in 5.9% (35) of episodes in reproductive women (i.e., between 15 and 50 years old) registered in the database. Among those, 16 suffered from comorbidities (3 asthma, 3 diabetes, 2 hypertension and 14 other comorbidities), and 14 complications occurred, mostly respiratory complications (7 episodes) and ENT complications (4 episodes). Only one pregnant woman stayed in the ICU. None of them received antiviral treatment for COVID-19, and no deaths were reported.
A total of 146 (4.0%) patients were employed in a healthcare facility: one (0.7%) was infected in a hospital and six (4.1%) were of unknown source.
By using an existing surveillance system for influenza and adapting it for the surveillance of SARS-CoV-2/COVID-19 cases, we quickly developed a large harmonised dataset of hospitalised COVID-19 cases in Switzerland. The data enable assessment of disease severity, risk factors, treatments and the clinical course of the disease. The data are useful for informing public health authorities and policy makers on the impact of health regulations put in place during the pandemic and has the potential to be used in modelling the future of the disease in the country. Due to the lack of data at the European level when the pandemic started in Europe, most models and measures taken against COVID-19 in Switzerland were based on Chinese data. The data obtained during this first wave will therefore enable healthcare workers and policy makers to respond more efficiently to the disease in the future.
Despite the system being rapidly set up, its use for real-time monitoring of the epidemic presented some challenges. Discussions with the FOPH on adapting the existing influenza database started in early February and the system was fully operational by the end of the same month, shortly after the first case was declared in Switzerland. This highlights the adaptability of the original system for surveillance of other respiratory viruses. Despite the strength of the system in place, it was difficult to obtain data within 48 hours of the diagnosis for most centres faced with a high number of cases. Some hospitals started the data collection later than others, and had to enter data both retrospectively and prospectively. Other centres were overwhelmed by the large number of cases and had to focus on patient care first, while at the same time trying to train new staff who could help with data entry. The overall median time difference between the first positive laboratory test and the inclusion in the database was 13 days (IQR 3–28). Figure 6 shows the time delay throughout the pandemic. The maximum delay was observed during the height of the epidemic in Switzerland, but quickly decreased as the number of cases decreased. Note that other Swiss entities also requested data from several participating hospitals, making it more difficult for them to report episodes within the required 48 hours.
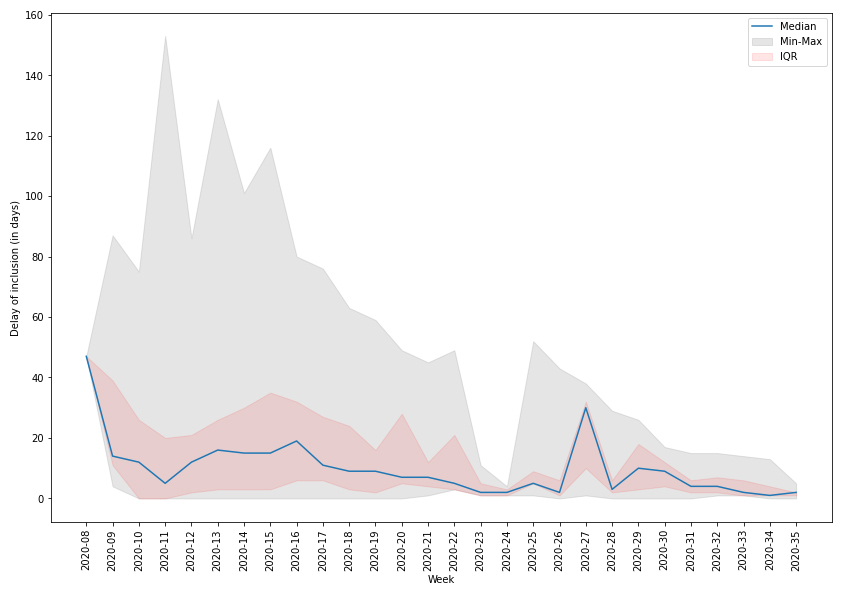
Figure 6 Time delay (in days) between the date of interest for the system and the inclusion of the patient in the database.
Using the same approach as the influenza database, data were checked weekly for inconsistencies and reported to the participating hospitals and the FOPH. Outliers in dates, impossible combinations of variables, length of stays and treatments, out of range numbers and missing values were consistently checked. This allowed the participating hospitals to correct wrong and missing data rapidly. Additional checks on the data, based on the patients’ records, were made by the supervising physicians in each hospital to ensure medical accuracy.
By including a large number of university and cantonal hospitals, we collected 3650 cases confirmed by PCR as of 1 September 2020 and 527 (14.7%) COVID-19 related deaths. On the same date, the mandatory reporting system put in place by the FOPH [14] had registered 4588 confirmed hospitalised patients related to COVID-19. Among these hospitalised patients, 945 (20.6%) died during their hospital stay. Overall, 59.9% of patients were male and 40.0% female. Comorbidities were observed in 3475 (75.7%) patients. Among thoese were hypertension in 60.8% of the patients, chronic cardiovascular diseases in 39.2%, diabetes in 27.4%, chronic respiratory diseases in 18.2%, oncological disease in 11.6% and immunosuppression in 4.9%. Those numbers match with the proportions found in the hospital database, except for hypertensive patients, cardiovascular pathologies and patients suffering from diabetes, for which the FOPH reporting system found higher proportions. The median age of the patients in the compulsory declaration system was also slightly higher than that in our database (70 versus 68 years old). Table 4 summarises the findings from the mandatory system.
Table 4 Characteristics of COVID-19/SARS-CoV-2 patients hospitalised up to 1 September 2020, in the mandatory reporting system of the Federal Office of Public Health.
|
Patients
(N = 4588) |
|||
|---|---|---|---|
| n | % | ||
| Gender |
Male | 2749 | 59.9 |
| Female | 1837 | 40.0 | |
| Other | 2 | 0.0 | |
| Age group |
<1 year | 2 | 0.0 |
| 1–5 | 7 | 0.1 | |
| 6–12 | 9 | 0.2 | |
| 13–19 | 44 | 1.0 | |
| 20–29 | 160 | 3.5 | |
| 30–39 | 179 | 3.9 | |
| 40–49 | 324 | 7.1 | |
| 50–59 | 583 | 12.7 | |
| 60–69 | 907 | 19.8 | |
| 70–79 | 1059 | 23.1 | |
| 80–89 | 979 | 21.3 | |
| 90–99 | 306 | 6.7 | |
| >100 | 7 | 0.1 | |
| Unknown | 22 | 0.5 | |
| Had at least one comorbidity |
Yes | 3475 | 75.7 |
| No | 625 | 13.6 | |
| Unknown | 488 | 10.6 | |
| – Diabetes |
Yes | 951 | 27.4 |
| No or Unknown | 2524 | 72.6 | |
| – Cardiovascular diseases |
Yes | 1361 | 39.2 |
| No or Unknown | 2114 | 60.8 | |
| – Hypertension |
Yes | 2112 | 60.8 |
| No or Unknown | 1363 | 39.2 | |
| – Respiratory diseases |
Yes | 633 | 18.2 |
| No or Unknown | 2842 | 81.8 | |
| – Oncological diseases |
Yes | 405 | 11.6 |
| No or Unknown | 3070 | 88.3 | |
| – Immunosuppressed |
Yes | 171 | 4.9 |
| No or Unknown | 3304 | 95.1 | |
| – Other |
Yes | 1608 | 46.3 |
| No or Unknown | 1867 | 53.7 | |
| Outcome |
Deceased | 945 | 20.6 |
| Alive | 3644 | 79.4 | |
Note: The age group <1year encompasses the <1 month and 1 month–5 years age group from our system, as the mandatory reporting system data contains age in years without decimals and no date of birth. Official data do not allow for separation between “No” and “Unknown”, hence their common category. Data courtesy of FOPH.
We nevertheless included approximatively 80% of the total number of SARS-CoV-2/COVID-19 hospitalised patients reported in the obligatory system of the FOPH up to 1 September 2020. The lower proportion of deaths in our system suggests that many patients may have died after discharge from the hospital, in another institution not captured by the system, or among patient hospitalised for less than 24 hours. We were also missing data on adult patients from three hospitals that only agreed to contribute paediatric cases, potentially explaining the difference in the median age in both systems.
The setting-up of this database is a first step towards an in-depth understanding of the SARS-CoV-2/COVID-19 epidemic in Switzerland, and is a powerful tool for the second wave, initially reported in China [15] and currently occurring in Europe. The data will allow authorities to tackle risk populations more precisely and adapt restrictions. The system was easily adapted from the influenza surveillance system in a matter of days by including and removing disease-specific information, which allowed a fast response in obtaining data. However, the lack of human resources, the severity of the pandemic and the multiplication of COVID-19 data collection in hospitals by Swiss medical societies and authorities made it difficult to obtain data rapidly. Therefore, timely monitoring of the epidemic was difficult, an issue that needs to be addressed rapidly for future pandemics by working more closely with officials to avoid overlapping demands.
Despite the drawback, of the database as a monitoring tool, the robust coverage of patients when compared to the mandatory reporting system makes it a powerful addition to the mandatory reporting system by adding more detailed information on the epidemiology, risk factors and clinical course of hospitalised patients, which is missing from the official system.
The surveillance is still ongoing and we plan, in the future, to include patients with negative PCR results but a suggestive clinical presentation combined with radiographic findings compatible with COVID-19, and patients with positive SARS-CoV-2 serology. We will distinguish these three modes of diagnosis: positive PCR ensures that the virus is present, whereas computed tomography and radiography show specific features of COVID-19 resulting in only a suspected diagnosis. Positive serology indicates that a patient was previously infected, and could therefore be of interest for hospitalisations due to complications of the infection (e.g., if the patient was not initially hospitalised).
There are ongoing discussions on linking patients from the database to their biological samples, in order to study associations between risk factors, complications and specific strains of the virus and, therefore, possibly establish microbiological surveillance of the disease.
Finally, we plan to study long-term consequences of infections, since the system allows us to track several hospital stays in affected patients.
The anonymised data can be accessed through a multi-stage process. Applicants must fill a concept-sheet (available in appendix 2) and send it to the team in charge of the study. An Executive Committee of experts and representatives of participating hospital will review the concept. Depending on the goal of the analysis, additional ethics clearance might be needed. Data will be restricted to the request and shared through a secure platform. All steps to access the data are described in the aforementioned concept-sheet, including contact details. Additional information can be found on the dedicated website: https://www.unige.ch/medecine/hospital-covid/.
The appendices are available for download as separate PDF files in the download section at https://doi.org/10.4414/smw.2021.20475.
The authors would like to thank all the participating centres’ teams, study nurses and physicians for their hard work and commitment to the study.
AT set up the system, including coding the database, checking for data inconsistencies, and wrote the scripts for analyses. He also drafted and corrected the present manuscript. AI helped set up the first version of the CRF, provided medical expertise and helped draft the first version of the manuscript. She is also the project leader for the Hôpitaux Universitaire de Genève. CB, LS, NT, AW, DF, PWS, MV, LD, MB, DVG, CK, AC, TR, YN, RG, UH, CB, FZ, SBS, NC, PZ, AU, and ANL were part of the data collection teams as local project leaders, provided feedback on the CRF and on each iteration of the manuscript. CG led the project for the Federal Office of Public Health. She helped design the CRF from an official perspective and provided administrative support. MR helped design the CRF and assess data quality throughout the data collection period. OK provided feedback and correction for each iteration of the manuscript.
This work was supported by the Swiss Federal Office of Public Health under reference 333.0-20/1. OK acknowledges additional support from the Swiss National Science Foundation (SNF) via grant #163878. PWS has received grant support from the Career funding program "Filling the Gap" of the Medical Faculty of the University of Zurich. With contributions of the Clinical Research Centre, Geneva University Hospitals and Faculty of Medicine, Geneva.
The authors acknowledges the following conflicts of interest: PWS has received travel grants from Gilead and Pfizer, and speakers honorary from Pfizer. UH is a member of the Coalition for Epidemic Preparedness Innovations (CEPI) Meta-Data Safety Monitoring Board, currently overseeing COVID-19 vaccine trials.
1Coronavirus Update (Live) [Internet] [cited 2020 Sept 4]. Available from: https://www.worldometers.info/coronavirus/
2 Zhu N , Zhang D , Wang W , Li X , Yang B , Song J , et al.; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–33. doi:.https://doi.org/10.1056/NEJMoa2001017
3World Health Organisation (WHO). nCoV 2019 situation (public) [Internet]. [cited 2020 Aug 12]. Available from: https://covid19.who.int
42019-nCoV outbreak is an emergency of international concern [Internet]. 2020 [cited 2020 June 24]. Available from: http://www.euro.who.int/en/health-topics/emergencies/pages/news/news/2020/01/2019-ncov-outbreak-is-an-emergency-of-international-concern
5World Health Organization (WHO). Novel Coronavirus (2019-nCoV) technical guidance: Early investigations [Internet]. [cited 2020 June 24]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/early-investigations
6Swiss Federal Office of Public Health (FOPH). Formulaires de déclaration [Internet]. [cited 2020 June 24]. Available from: https://www.bag.admin.ch/dam/bag/fr/dokumente/mt/msys/covid-19-meldeformular-hospitalisierte.pdf.download.pdf/OFSP_covid19_formulaire-de-declaration_patients-hospitalises.pdf
7World Health Organization (WHO). Coronavirus disease (COVID-19) outbreak - Situation dashboard for Europe [Internet]. [cited 2020 Sept 4]. Available from: https://who.maps.arcgis.com/apps/opsdashboard/index.html#/ead3c6475654481ca51c248d52ab9c61
8Thiabaud A, Iten A, Troillet N, Senn L, Flury D, Kuster SP, et al. Hospital-based surveillance of influenza in Switzerland: a pilot study – season 2018/19. Under review.
9Project REDCap [Internet]. [Accessed 2020 Aug 25]. Available from: https://www.project-redcap.org
10World Health Organization (WHO). COVID-19 core case report form acute respiratory infection clinical characterisation data tool [Internet]. [cited 2020 June 24]. Available from: https://media.tghn.org/medialibrary/2020/06/ISARIC_WHO_nCoV_CORE_CRF__Modules.pdf
11 Lim WS , van der Eerden MM , Laing R , Boersma WG , Karalus N , Town GI , et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58(5):377–82. doi:.https://doi.org/10.1136/thorax.58.5.377
12Swissnoso - Prevention & control of healthcare-associated COVID-19 outbreaks, [internet]. [cited 2020 November 27] https://www.swissnoso.ch/fileadmin/swissnoso/Dokumente/5_Forschung_und_Entwicklung/6_Aktuelle_Erreignisse/200515_Prevention_and_control_of_healthcare-associated_COVID-19_outbreaks_V1.0_ENG.pdf
13 Charlson M , Szatrowski TP , Peterson J , Gold J . Validation of a combined comorbidity index. J Clin Epidemiol. 1994;47(11):1245–51. doi:.https://doi.org/10.1016/0895-4356(94)90129-5
14Swiss Federal Office of Public Health (FOPH). New coronavirus: Situation in Switzerland [Internet]. [cited 2020 Aug 12]. Available from: https://www.bag.admin.ch/bag/en/home/krankheiten/ausbrueche-epidemien-pandemien/aktuelle-ausbrueche-epidemien/novel-cov/situation-schweiz-und-international.html
15World Health Organisation (WHO). A cluster of COVID-19 in Beijing, People’s Republic of China[Internet]. [cited 2020 June 24]. Available from: https://www.who.int/news-room/detail/13-06-2020-a-cluster-of-covid-19-in-beijing-people-s-republic-of-china
This work was supported by the Swiss Federal Office of Public Health under reference 333.0-20/1. OK acknowledges additional support from the Swiss National Science Foundation (SNF) via grant #163878. PWS has received grant support from the Career funding program "Filling the Gap" of the Medical Faculty of the University of Zurich. With contributions of the Clinical Research Centre, Geneva University Hospitals and Faculty of Medicine, Geneva.
The authors acknowledges the following conflicts of interest: PWS has received travel grants from Gilead and Pfizer, and speakers honorary from Pfizer. UH is a member of the Coalition for Epidemic Preparedness Innovations (CEPI) Meta-Data Safety Monitoring Board, currently overseeing COVID-19 vaccine trials.