Autophagy and coronavirus infection – a Trojan horse or Achilles heel?
DOI: https://doi.org/10.4414/smw.2021.20468
Kristina
Seilerab, Mario P.
Tschanab
a Institute of Pathology, University of Bern, Switzerland
b Graduate School for Cellular and Biomedical Sciences, University of Bern, Switzerland
The year 2020 introduced far-reaching changes into the world’s daily routine after the emergence of a coronavirus (SARS-CoV-2), which quickly spread to cause a global pandemic, infecting almost 70 million people and claiming more than 1.5 million lives around the globe as of early December 2020. Huge efforts are currently being undertaken by science, industry and governments together to identify drugs, vaccines and transmission prevention strategies in order to keep the spread of the virus under control and manage health, economic and social damage.
In an effort to combat SARS-Cov-2, clinicians and scientists are repurposing known drugs to accelerate compound identification and speed up the approval process. One of the first compounds reported to potentially have a beneficial effect on viral load and patient mortality was chloroquine (reviewed in [1]). However, clinical trials during the early stages of the disease spread were done rapidly and often without proper controls, leading to conflicting results.
Chloroquine, or the closely related hydroxychloroquine, is a widely available drug with well-known pharmacokinetics. It rapidly diffuses across membranes and accumulates in the lower pH environment of lysosomes, where it becomes protonated and trapped. Accumulation of the compound within lysosomes raises lysosomal pH and inhibits fusion between endosomes and lysosomes, and therefore has a major impact on lysosomal function and impedes late stages of autophagy where autophagosomes fuse with lysosomes [2]. The compound has been shown to be effective against a broad range of viruses in vitro, hampering crucial steps of low pH-dependent viral replication [3, 4]. Chloroquine also inhibits terminal glycosylation of the angiotensin converting enzyme-2 (ACE2) receptor through which SARS-CoV-2 enters the cell [5] (fig. 1). In vitro, non-glycosylated ACE2 interacts less efficiently with the spike protein of SARS-CoV2. It is therefore not entirely clear whether chloroquine’s effect on late autophagy is necessarily the cause of its general antiviral activity in vitro. In the case of COVID-19, the US Food and Drug Administration (FDA) revoked the Emergency Use Authorization for hydroxychloroquine in the treatment of COVID-19 after interim analysis of two large randomised trials showed no mortality benefit [6–8]. A recent meta-analysis, preprinted in medRxiv, by a team from the University of Basel compared data of 26 clinical trials investigating the use of hydroxychloroquine. While this publication still lacks peer-review, the meta-analysis found no survival benefit for patients receiving hydroxychloroquine [9].
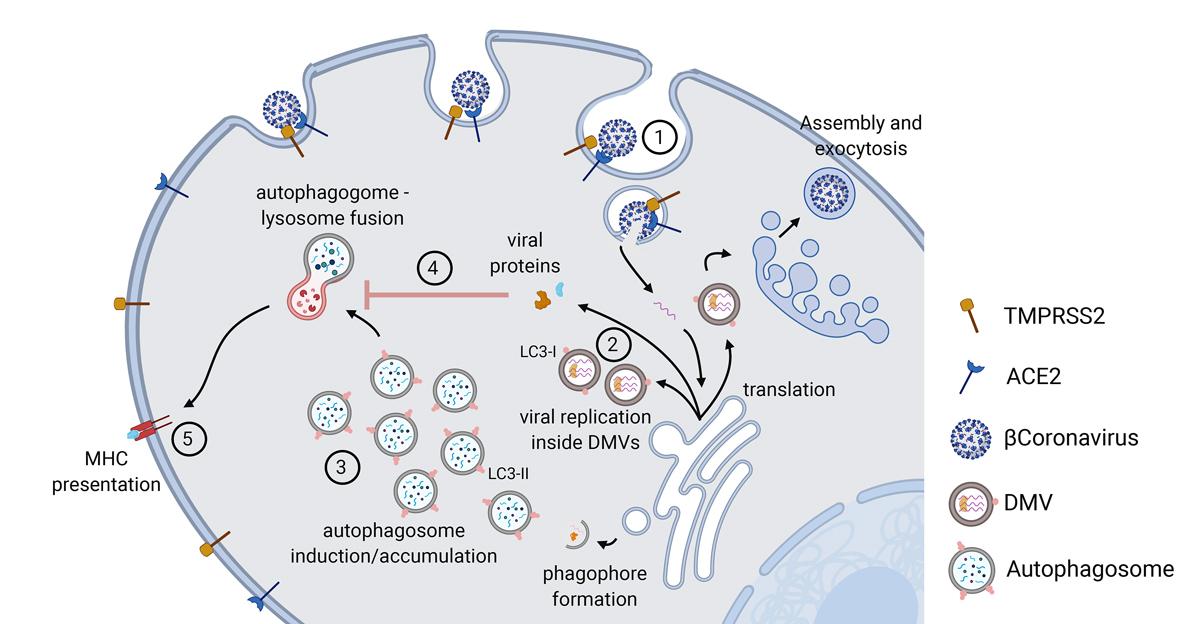
Figure 1
Overview of the autophagy pathway during viral infection. 1. ACE2/TMPRSS2 mediated endocytosis. 2. Replication of virus inside ER-derived DMVs containing LC3-I. 3. Induction of early stages of autophagy and accumulation of autophagosomes. 4. Viral proteins block the fusion event between autophagosomes and lysosomes, thereby blocking late stages of autophagy. 5. MHC-mediated presentation of viral particles upon autophagic degradation. Created with BioRender.com.
ACE2 = angiotensin converting enzyme-2; DMV = double membraned vesicles; ER = endoplasmic reticulum; LC3 = microtubule-associated protein 1A/1B-light chain 3B; MHC = major histocompatibility complex; TMPRSS2 = a type II transmembrane serine protease
Early on during the first wave of infections in April, a group from the virology department at Berlin’s Charité showed great potential for autophagy modulators, including spermidine, Akt (protein kinase B) inhibitor MK-2206 and Beclin-1 stabilising niclosamide, to inhibit SARS-CoV-2 propagation (preprint, not peer-reviewed) [10]. Many of the more promising compounds currently investigated in clinical trials are indeed autophagy modulators, thus substantiating the need for an understanding of how coronaviruses interact with and utilise components of the autophagy pathway.
Autophagy is a multistep process for lysosomal degradation of cytoplasmic content such as damaged organelles, protein aggregates or long-lived proteins, and hence ensures a supply of biomass in times of nutrient starvation and stress. Although there are different subtypes of autophagy depending on cargo recognition and the method of delivery to the lysosome, we focus solely on macroautophagy for the purposes of this article. Macroautophagy (hereafter autophagy) traps cytoplasmic contents through the formation of a thin membrane around the cargo, called the phagophore, which furthermore expands and closes to become the double-membraned autophagosome.
Autophagy is involved in the immune response through clearance of intracellular pathogens (xenophagy/virophagy), as well as through processing of antigens for major histocompatibility complex (MHC)-dependent presentation to T cells. Several viruses have been reported to hijack the cellular autophagy machinery to provide scaffold functions for replication purposes (reviewed in [11]). Beta coronaviruses (bCov), the family to which SARS-CoV2 belongs, have been shown to induce an autophagy-related gene 5 (ATG5)-dependent upregulation of autophagosome formation, but subsequently inhibit their maturation [12, 13]. Electron microscopy of cells infected with mouse hepatitis virus (MHV), a bCov family member used to study bCovs in the laboratory, revealed that viral replication is indeed localised within double membraned vesicles (DMVs, fig. 1, caption 2), much like autophagosomes [14]. However, the same authors as well as others found that those DMVs contain LC3-I, rather than LC3-II, which is found on autophagosomes, and that DMVs occur after infection even in an autophagy-deficient background [14] (LC3 = microtubule-associated protein 1A/1B-light chain 3B). The upregulation of LC3-I containing DMVs after bCov infection could therefore indicate viral hijacking of another pathway, endoplasmic reticulum (ER)-associated degradation (ERAD), as DMV membranes are thought to be derived from the ER [15]. It is not entirely clear if or how viral replication inside DMVs is connected to autophagy, but there is evidence of an increasing number of autophagosomes (fig. 1, caption 3) upon bCov infection [12, 14] and, importantly, an up to 86% reduction of MHV replication in autophagy-incompetent cells [16]. In line with these findings, Prentice et al. showed an upregulation of autophagy upon MHV infection by looking at degradation of long-lived proteins as an autophagic flux assay [16]. Assessment of autophagy and autophagic flux is generally challenging, as it is a multistep dynamic process, which can involve different protein complexes and does not always include lipidation of LC3B [17]. We would therefore like to stress the importance of assessing autophagic activity by more than one method [18]. Autophagy is a complex process requiring a multitude of proteins with substantial redundancy; hence, if looking closely, one finds conflicting results with regards to viral replication and autophagy inhibition. It is therefore believed that, although targeting a single isolated autophagy gene may not always disrupt the autophagic machinery as a whole, there may very well be unconventional functions of ATGs important for the viral life cycle. This notion is supported by an ATG-proteome specific small interfering RNA (siRNA) screen across cells infected with six different viruses, where knockdown of autophagy proteins uncovered a complex landscape of both viral suppression and promotion, depending on the target [19]. Interestingly, for MHV infection, knocking down multiple redundant components of the autophagy machinery almost always led to a decrease in viral replication.
For viruses, positioning themselves within a degradation-competent vesicle and inducing autophagy has the advantage of maintaining high levels of ATP needed for replication. On the other hand, upregulation of autophagy upon bCov infection could potentially reduce virulence through increased clearing of viral particles. Certain coronaviruses seem to have addressed this by developing strategies to downregulate Beclin1 and thus inhibit late-stage autophagy (reviewed in [20]). Beclin1 is also important for early membrane rearrangements during autophagosome formation [21], but increased numbers of autophagosomes without an increase in autolysosomes [22] upon bCov infection points towards an inhibition of mostly late-stage autophagy, as Beclin1 is crucial for fusion of autophagosomes, and potentially DMVs, with the lysosome [22] (fig.1 caption 4). Viral targeting of Beclin1 is also interesting as Beclin1 is a key player in antiviral interferon signalling [23]. For SARS-CoV, it has been shown that the presence of a SARS-CoV specific protease led to Beclin1 associating with the stimulator of interferon genes pathway (STING) and resulted in a decreased interferon-β response [13]. Whereas massive interferon signalling arguably aggravates multi-organ damage in severely ill patients, an adequate interferon response at an early stage of infection may well prove beneficial. This is highlighted by most recent genome-wide studies identifying certain genetic determinants of severe disease that are in part connected to interferon signalling [24].
There is a large body of literature on autophagy and its role in viral replication. Although it certainly is crucial in the life cycles of most, if not all, viruses, whether the autophagy machinery is hijacked to benefit viral replication, or whether it is promoting antiviral defence through clearance of viral particles and antigen presentation seems very much to depend on the specific type of virus. In the case of SARS-Cov-2, the literature at this point would almost suggest a dual role of promoting viral replication through the early stages of autophagy and blocking later stages to prevent clearance of viral particles and/or antigen presentation (fig. 1, caption 5). The evidence so far cautiously points towards an enhanced clearing of SARS-COv-2 infection through autophagy-inducing agents. However, to date there is no direct experimental evidence to pin down autophagy as either a pro- or antiviral process during COVID-19 disease. Whether or not autophagy modulators will eventually be useful in the treatment of COVID-19 might also depend on a patient’s individual autophagic status, and stratification might therefore be of importance when assessing the efficiency of these drugs. Further experimental investigations, as well as analysis of autophagic activity in patient samples, will hopefully advance treatment options and lead to a better understanding of how different stages of autophagy are involved in viral replication and infection control.
Acknowledgements
The support of the TRANSAUTOPHAGY COST Action CA15138 and Life Sciences Switzerland, Section Autophagy is highly appreciated.
References
1
Colson
P
,
Rolain
JM
,
Raoult
D
. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents. 2020;55(3):105923. doi:.https://doi.org/10.1016/j.ijantimicag.2020.105923
2
Solomon
VR
,
Lee
H
. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625(1-3):220–33. doi:.https://doi.org/10.1016/j.ejphar.2009.06.063
3
Savarino
A
,
Di Trani
L
,
Donatelli
I
,
Cauda
R
,
Cassone
A
. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6(2):67–9. doi:.https://doi.org/10.1016/S1473-3099(06)70361-9
4
Rolain
JM
,
Colson
P
,
Raoult
D
. Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infections in the 21st century. Int J Antimicrob Agents. 2007;30(4):297–308. doi:.https://doi.org/10.1016/j.ijantimicag.2007.05.015
5
Vincent
MJ
,
Bergeron
E
,
Benjannet
S
,
Erickson
BR
,
Rollin
PE
,
Ksiazek
TG
, et al.
Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2(1):69. doi:.https://doi.org/10.1186/1743-422X-2-69
6Office of the Commissioner. Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine. U.S. Food and Drug Administration. Available from: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and (2020)
7WHO discontinues hydroxychloroquine and lopinavir/ritonavir treatment arms for COVID-19. Available from: https://www.who.int/news-room/detail/04-07-2020-who-discontinues-hydroxychloroquine-and-lopinavir-ritonavir-treatment-arms-for-covid-19
8
Horby
P
,
Mafham
M
,
Linsell
L
,
Bell
JL
,
Staplin
N
,
Emberson
JR
, et al., RECOVERY Collaborative Group. Effect of Hydroxychloroquine in Hospitalized Patients with Covid-19. N Engl J Med. 2020;383(21):2030–40. doi:.https://doi.org/10.1056/NEJMoa2022926
9
Axfors
C
,
Schmitt
AM
,
Janiaud
P
,
van ‘t Hooft
J
,
Abd-Elsalam
S
,
Abdo
EF
et al.
Mortality outcomes with hydroxychloroquine and chloroquine in COVID-19: an international collaborative meta-analysis of randomized trials. medRxiv. 2020;9.16:20194571. doi:
10
Gassen
NC
,
Papies
J
,
Bajaj
T
,
Dethloff
F
,
Emanuel
J
,
Weckmann
K
,
Heinz
DE
,
Heinemann
N
,
Lennarz
M
,
Richter
A
,
Niemeyer
D
,
Corman
VM
,
Giavalisco
P
,
Drosten
C
,
Müller
MA
. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. bioRxiv. 2020;04.15:997254. doi:
11
Kudchodkar
SB
,
Levine
B
. Viruses and autophagy. Rev Med Virol. 2009;19(6):359–78. doi:.https://doi.org/10.1002/rmv.630
12
Cottam
EM
,
Whelband
MC
,
Wileman
T
. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426–41. doi:.https://doi.org/10.4161/auto.29309
13
Chen
X
,
Wang
K
,
Xing
Y
,
Tu
J
,
Yang
X
,
Zhao
Q
, et al.
Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014;5(12):912–27. doi:.https://doi.org/10.1007/s13238-014-0104-6
14
Reggiori
F
,
Monastyrska
I
,
Verheije
MH
,
Calì
T
,
Ulasli
M
,
Bianchi
S
, et al.
Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7(6):500–8. doi:.https://doi.org/10.1016/j.chom.2010.05.013
15
Knoops
K
,
Kikkert
M
,
Worm
SH
,
Zevenhoven-Dobbe
JC
,
van der Meer
Y
,
Koster
AJ
, et al.
SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. doi:.https://doi.org/10.1371/journal.pbio.0060226
16
Prentice
E
,
Jerome
WG
,
Yoshimori
T
,
Mizushima
N
,
Denison
MR
. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279(11):10136–41. doi:.https://doi.org/10.1074/jbc.M306124200
17
Noda
T
,
Kageyama
S
,
Fujita
N
,
Yoshimori
T
. Three-Axis Model for Atg Recruitment in Autophagy against Salmonella. Int J Cell Biol. 2012;2012:389562. doi:.https://doi.org/10.1155/2012/389562
18
Klionsky
DJ
,
Abdelmohsen
K
,
Abe
A
,
Abedin
MJ
,
Abeliovich
H
,
Acevedo Arozena
A
, et al.
Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. doi:.https://doi.org/10.1080/15548627.2015.1100356
19
Mauthe
M
,
Langereis
M
,
Jung
J
,
Zhou
X
,
Jones
A
,
Omta
W
, et al.
An siRNA screen for ATG protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. J Cell Biol. 2016;214(5):619–35. doi:.https://doi.org/10.1083/jcb.201602046
20
Miller
K
,
McGrath
ME
,
Hu
Z
,
Ariannejad
S
,
Weston
S
,
Frieman
M
, et al.
Coronavirus interactions with the cellular autophagy machinery. Autophagy. 2020;16(12):2131–9. doi:.https://doi.org/10.1080/15548627.2020.1817280
21
Itakura
E
,
Kishi
C
,
Inoue
K
,
Mizushima
N
. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–72. doi:.https://doi.org/10.1091/mbc.e08-01-0080
22
Gassen
NC
,
Niemeyer
D
,
Muth
D
,
Corman
VM
,
Martinelli
S
,
Gassen
A
, et al.
SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat Commun. 2019;10(1):5770. doi:.https://doi.org/10.1038/s41467-019-13659-4
23
Wirawan
E
,
Lippens
S
,
Vanden Berghe
T
,
Romagnoli
A
,
Fimia
GM
,
Piacentini
M
, et al.
Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8(1):6–17. doi:.https://doi.org/10.4161/auto.8.1.16645
24
Ellinghaus
D
,
Degenhardt
F
,
Bujanda
L
,
Buti
M
,
Albillos
A
,
Invernizzi
P
, et al., Severe Covid-19 GWAS Group. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med. 2020;383(16):1522–34. doi:.https://doi.org/10.1056/NEJMoa2020283
