
Figure 1 Domain structure of murine DNA methyltransferases. ADD = ATRX-DNMT3-DNMT3L; BAH = bromo-associated homology; MTase = methyltransferase; RFTS = replication foci targeting sequence
DOI: https://doi.org/10.4414/smw.2020.20329
While the field of genetics studies heredity, in which genes that are encoded in the DNA represent the basic units of inheritance, the field of epigenetics (from the Greek ἐπί, meaning “above”) studies those mechanisms that effectuate heritable changes without altering the underlying DNA sequence itself. Such changes are known as epigenetic modifications and exist in a variety of forms. The only epigenetic modification that takes place directly at the level of the nucleosides is DNA methylation, which comprises the coupling of a methyl group (CH3) to DNA bases. Although other contexts are permissive, most methylation in vertebrates appears on the 5′ position of the pyrimidine ring of cytosine, resulting in 5-methylcytosine (5meC) [1]. Apart from a low prevalence of 5meC at CpA, CpT, and CpC sequences in certain cell types, mammalian cytosine methylation is generally found in the setting of a cytosine followed by a guanine on the same strand, constituting a self-complementary CpG dinucleotide [2, 3]. Due to the inherent mutability of 5meC the general occurrence of CpGs in the genome is low, with the exception of so-called CpG islands (CGIs), which are short regions of high CpG content that are commonly associated with gene promoters [4]. Whereas approximately 70% of CpG sites in the adult vertebrate genome are methylated, the majority of CGIs remain free from methylation [4]. CGI promoter methylation can regulate transcription of associated genes in cis, which is particularly important for X chromosome inactivation, genomic imprinting, and germline-specific genes [5–7]. In addition, DNA methylation has been shown to control the silencing of transposable elements [8]. Together, this demonstrates that cytosine methylation has a central regulatory function in a variety of biological processes.
DNA methylation is a dynamic process, which can be divided into three stages: establishment, maintenance, and removal. The DNA methyltransferase enzymes DNMT3A and DNMT3B are the main proteins responsible for de novo methylation; in addition, DNMT3C has recently been reported to be a rodent-specific methyltransferase that is involved in silencing young retrotransposons specifically in the male germline [9], and DNMT3L lacks a functional catalytic domain but serves as an important cofactor for DNMT3A and DNMT3B [10]. Representing a different branch of the DNMT family, DNMT1 is seen as the main maintenance methyltransferase, which after replication is recruited to hemimethylated DNA to ensure faithful propagation of methylation patterns [11]. When the activity of DNMTs is impaired, passive DNA methylation leads to gradual loss of 5meC over several cell divisions. Alternatively, ten-eleven translocation (TET) methylcytosine dioxygenases can initiate active DNA demethylation by catalysing the oxidation of 5meC into 5-hydroxymethylcytosine (5hmC), which subsequently can be processed into unmethylated cytosine by various means [12, 13]. DNMTs and TETs are hence the main regulators of the dynamics of DNA methylation [14, 15].
While promoter methylation has typically been associated with gene silencing, methylation at gene bodies is in contrast positively correlated with transcription, suggesting that the regulation and function of DNA methylation is largely context-dependent [2, 16]. Given the global distribution of DNA methylation along the genome, local context is expected to strongly influence how this mark regulates various biological processes. This context is for a major part defined by another class of epigenetic modifications, in the form of histone marks. DNA is wrapped around an octamer of histone proteins (two of each histone H2A, H2B, H3, and H4), of which both the core and tail domains contain a large number of amino acid residues that can be post-translationally modified. The most important and frequently occurring modifications on histones are methylation and acetylation [17]. Depending on their properties, different histone modifications, alone or in combination, could have their own specific effects on transcription, leading to the concept that a potential “histone code” could provide regulatory cues for gene expression. Moreover, numerous studies have investigated the connection between histone marks and DNA methylation, revealing an intriguing interplay [18].
In this review, we will address the crosstalk between DNA methylation and other epigenetic modifications, with a focus on how histone modifications influence recruitment of the different DNMT family members to the DNA. In addition, we will describe several human disease phenotypes that have been associated with aberrant DNMT recruitment, and what these diseases teach us about the molecular mechanisms underlying their specific binding.
The first member of the DNA methyltransferase family to be cloned and sequenced in eukaryotes was named DNMT1 [19]. DNMT1 is a multi-domain protein consisting of, from N- to C-terminus, a replication foci targeting sequence (RFTS), a zinc finger CXXC domain, two tandemly connected bromo-associated homology (BAH) domains, and the catalytic methyltransferase (MTase) domain [20] (fig. 1). Furthermore, its non-catalytic N-terminus has been revealed to mediate protein-protein interactions [21]. The preferred substrate for this enzyme was demonstrated to be hemimethylated DNA, suggesting that it could facilitate the semi-conservative inheritance of methyl moieties during DNA replication [22]. Although DNMT1 is the only DNA methyltransferase that harbours this substrate specificity and has therefore been designated as the maintenance DNMT, it in fact also contains considerable de novo methylation activity both in vitro and in vivo [23, 24]. Vice versa, in certain cell types the de novo methyltransferases DNMT3A and DNMT3B have been shown to contribute to maintenance methylation as well [25, 26]. While the classical ‘division of labour’ model of DNMT1 operating in maintenance and the DNMT3s in de novo methylation still holds true for certain developmental contexts, it generally is thus an oversimplified, but nevertheless useful framework until all its underlying principles are understood [27, 28].

Figure 1 Domain structure of murine DNA methyltransferases. ADD = ATRX-DNMT3-DNMT3L; BAH = bromo-associated homology; MTase = methyltransferase; RFTS = replication foci targeting sequence
How DNMT1 is specifically recruited to hemimethylated DNA has been extensively studied, revealing a complex mechanism involving several factors. Apart from its inherent preference for hemimethylated cytosine, it was discovered early on that DNMT1 interacts with the DNA clamp proliferating cell nuclear antigen (PCNA), which localises at the replication fork and thereby ensures DNMT1 proximity to newly synthesised DNA [29]. However, the later finding that PCNA is not strictly required for this process indicated that other players must be involved [30]. The most important player was subsequently identified to be the E3 ubiquitin-protein ligase ubiquitin-like with PHD and RING finger domains 1 (UHRF1), knockout of which in mice mimics the Dnmt1-/- phenotype [31]. UHRF1 binds to hemimethylated CpG sites via its SET and RING associated (SRA) domain by a base-flipping mechanism, and engages DNMT1 through a direct protein-protein interaction between its own ubiquitin-like (UBL) domain and the RFTS of DNMT1 [32, 33]. In addition, UHRF1 binds to unmodified arginine 2 on histone H3 (H3R2) and to di- and tri-methylated lysine 9 on histone H3 (H3K9me2/3) via its plant homeodomain finger (PHD) and tandem tudor domain (TTD), respectively, thereby contributing to DNMT1 recruitment to different chromatin environments [34–36]. Recent work suggested also a direct interaction between H3K9me3 and the RFTS domain of DNMT1, which would further reinforce the association between this histone mark and maintenance methylation [37].
Adding another layer of epigenetic complexity, UHRF1 catalyses mono-ubiquitinylation of H3 at lysines 14, 18, and 23 [38]. These residues are then bound by the RFTS domain of DNMT1, which is important for both its recruitment as well as its methyltransferase activity [38]. Indeed, binding of the RFTS to ubiquitylated H3 tails releases its auto-inhibitory binding to the MTase domain, leading to enhanced methylation [20]. Recently, an additional ubiquitin-dependent mechanism for DNMT1 recruitment was proposed, in which UHRF1 ubiquitylates PCNA-associated factor 15 (PAF15) on chromatin in a DNA replication-coupled manner [39]. In a mode of action similar to PCNA, PAF15-Ub2 specifically binds to replicating chromatin and forms a complex with DNMT1, thereby contributing to stable maintenance of DNA methylation [39].
In contrast to DNMT1, the DNMT3 branch of the DNMT family consists of several closely related members, which in terms of structure represent different variations on the same theme (fig. 1). DNMT3A and DNMT3B are the largest proteins and are built up of a relatively long N-terminal region devoid of any known domains, followed by three chromatin-interacting domains. From N- to C-terminus these are the proline-tryptophan-tryptophan-proline (PWWP) domain, the ATRX-DNMT3-DNMT3L (ADD) domain, and the MTase domain. The rodent-specific DNMT3C enzyme arose through duplication of the Dnmt3b gene, but during evolution lost its PWWP domain [40]. DNMT3L likewise lost its PWWP domain, and additionally lacks a functional MTase domain, making it the shortest family member [41]. Although DNMT3L does not possess any inherent DNA methyltransferase ability, it serves as a cofactor for DNMT3A and DNMT3B by directly interacting with these proteins and stimulating their activity, which is particularly important for the establishment and expression of maternally imprinted genes [10, 42, 43].
DNMT3A and DNMT3B are the main enzymes responsible for setting DNA methylation without pre-existing methyl groups being present as a template, more commonly known as de novo methylation [44]. Despite their similarity and partial redundancy, neither DNMT3A nor DNMT3B can fully compensate for the loss of the other enzyme, indicating their distinctive functionalities [44]. Although it had been known for a long time that DNMT3A and DNMT3B possess intrinsic affinity for CpG sites, structural insights into this observation were only recently obtained [45–49]. These studies present crystal structures of the catalytic domains of human DNMT3A or DNMT3B in complex with DNMT3L, providing the structural basis for CpG-specific de novo DNA methylation.
Furthermore, the observation that DNA methylation is not evenly distributed over the genome suggests that CpG binding by the MTase domain is not the only defining factor for DNMT3 occupancy. Indeed, with the ADD and PWWP domains DNMT3A and DNMT3B possess two chromatin-interacting domains that must be taken into account in this regard. The first indications of the function of the ADD domain came from peptide interaction assays that showed that DNMT3L through its N-terminal region specifically interacts with the tail of histone H3, and that this interaction is abrogated by methylation of H3K4 [50]. That binding to unmethylated H3K4 is a core function of the ADD domain that is conserved across the DNMT3 family was subsequently confirmed by studies with DNMT3A and DNMT3B [51–53]. Moreover, this finding provided an explanation for the previously described anticorrelation between histone H3K4 methylation and DNA methylation [54, 55]. As shown for DNMT3A, binding to H3K4me0 by the ADD domain does not only regulate recruitment, but also stimulates methyltransferase activity through releasing an autoinhibitory interaction with the MTase domain [56].
Yet H3K4me3 is not the only histone modification involved in DNMT3 recruitment. Using peptide arrays, it was shown that the PWWP domain of DNMT3A specifically recognises H3K36me3 [57]. A recent study used chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) and found DNMT3A in vivo to be predominantly localised in H3K36me2 and H3K36me3 domains, with preferential enrichment in regions with high H3K36me2 compared to H3K36me3 [58]. This finding was recapitulated in vitro, with the DNMT3A-PWWP domain recognising both H3K36me2 and H3K36me3, but exhibiting greater affinity for H3K36me2 [58]. Similar in vivo experiments had already uncovered the targeted recruitment of DNMT3B to H3K36me3-decorated bodies of transcribed genes [59]. Since both DNMT3C and DNMT3L lost their PWWP domain, H3K36 methylation-mediated recruitment seems to be specific for DNMT3A and DNMT3B, although these could in turn recruit the other DNMTs through protein-protein interactions [40, 41]. In line with these findings, genome-wide profiling has established a positive correlation between H3K36me2/3 and DNA methylation [58].
Intriguingly, even though H3K36 methylation has been found to negatively correlate with the repressive histone mark H3K27me3, the longer isoform of DNMT3A (DNMT3A1) was discovered to localise to the H3K27me3-positive shores of CGIs [60, 61]. Apart from adding another histone mark to the code defining DNMT3 binding, it also suggests that more isoform-specific binding patterns may exist for DNMTs, since this preferential localisation was not found for the shorter isoform DNMT3A2 [60]. The exact relationship between DNA methylation and H3K27me3 has as yet remained elusive, but genome-wide assessments point to an anticorrelation, and indeed H3K27me3-enriched CGIs have been found to be devoid of DNA methylation [58, 62]. Rather than a direct recruiting function for H3K27me3, this implies recruitment of DNMT3A1 through indirect means, the underlying mechanisms of which must still be further elucidated.
Other mechanisms of DNMT3 recruitment have additionally been described. For example, the histone methyltransferase G9a has been demonstrated to recruit DNMT3A and DNMT3B through a direct interaction with their MTase domains, thereby connecting H3K9 methylation and de novo DNA methylation [63]. Similarly, the H3K27 histone methyltransferase EZH2 has been reported to interact with DNMT1 as well as DNMT3A/B and DNMT3L [64, 65], although this recruitment was later shown not to be sufficient for de novo DNA methylation and a direct role for Polycomb group proteins in regulating DNA methylation remains enigmatic [66–68]. Further protein-protein interactions between DNMT3s and the histone methyltransferases SUV39H1 and SETDB1 have also been described, suggesting that the link between histone and DNA methylation may be a more general one [69, 70]. However, it remains to be determined to what extent such connections would be causative instead of merely correlative.
All in all, recruitment of DNMTs is an intricate process in which the different layers are tightly interwoven. The robustness ensuing from this multi-layered complexity points to an imperative function for proper DNMT localisation in healthy individuals; in line with this, mouse embryos homozygous for knockout of the Dnmt1 or Dnmt3b gene die before birth, while Dnmt3a-/- mutant mice become runted and die approximately four weeks after birth [44, 71]. Nevertheless, a few diseases in humans have been described that could be caused by aberrant DNMT recruitment.
Mutations in the RFTS domain of DNMT1 have been associated with two different kinds of neurological disorders. The first one of these is hereditary sensory and autonomic neuropathy type 1 (HSAN1) with dementia and hearing loss, a form of neurodegeneration [72] (table 1). The identified mutations in DNMT1 were shown to result in protein misfolding, decreased enzymatic activity, and impaired heterochromatin binding, leading to the disease phenotype. On the level of the methylome, this translates to moderate global hypomethylation of cytosines but local hypermethylation at CGIs [72]. The second disease caused by mutations in the DNMT1-RFTS domain is autosomal dominant cerebellar ataxia, deafness and narcolepsy (ADCA-DN) [73] (table 1). Interestingly, both conditions are characterised by a late onset, proposing an important role for correct DNA maintenance methylation recruitment in adult neurons.
Table 1 Diseases associated with aberrant DNMT recruitment.
| Disease | Gene | Mutations | Molecular effect | References |
|---|---|---|---|---|
| HSAN1 (hereditary sensory and autonomic neuropathy type 1 with dementia and hearing loss) | DNMT1 | Y495C D490E-P491Y |
Protein misfolding, decreased enzymatic activity, and impaired heterochromatin binding | [72] |
| ADCA-DN (autosomal dominant cerebellar ataxia, deafness and narcolepsy) | DNMT1 | A570V G605A V606F |
Not tested, but hypothesised altered DNA binding and/or protein-protein interactions | [73] |
| Various haematological malignancies (AML, T-ALL, CMML, MDS, ARCH) | DNMT3A | Hotspot at R882; other disease-causing mutations over the entire length of the gene | Reduced methyltransferase activity, decreased or enhanced DNA binding, impaired ability to multimerise on the DNA, etc. | [74–77] |
| Tatton-Brown-Rahman syndrome | DNMT3A | Several in the PWWP, ADD, and MTase domains, e.g. I310N M548K R749C |
Not tested, but hypothesised altered binding to histone modifications and/or domain-domain interactions | [78] |
| Growth retardations | DNMT3A | W330R | Abrogated H3K36me2/3 interaction | [79] |
| D333N | [80] | |||
| Paraganglioma | DNMT3A | K299I R318W |
Not tested, but hypothesised abrogated H3K36me2/3 interaction | [81] |
| ICF syndrome (immunodeficiency, centromeric instability, and facial anomalies) | DNMT3B | Several in both the MTase and PWWP domain, e.g. S282P | Abrogated H3K36me3 interaction | [59, 82–84] |
Regarding the DNMT3 family, aberrant recruitment of mostly DNMT3A has been implicated in disease. Mutations in the DNMT3A gene have been widely associated with various haematological malignancies, most frequently with acute myeloid leukaemia (AML), but also including T-cell acute lymphoblastic leukaemia (T-ALL), chronic myelomonocytic leukaemia (CMML), and myelodysplastic syndrome (MDS) [74, 75] (table 1). In addition, DNMT3A mutations have been found to cause age-related clonal haematopoiesis (ARCH), which has been associated with not only haematological but also cardiovascular disease [76, 77] (table 1). Although one clear mutational hotspot exists at R882 in the MTase domain, disease-causing mutations have been found to occur over the entire length of the gene, leading to reduced methyltransferase activity, but also decreased or enhanced DNA binding, or impaired ability to multimerise on the DNA. The exact mechanisms by which these mutations drive haematological disease remain to be clarified but are often associated with genome-wide changes in the DNA methylation landscape. Through as yet unknown pathways this influences the cells’ ability to switch from self-renewal to differentiation, resulting in various kinds of neoplasms [74].
Furthermore, DNMT3A mutations have been linked to abnormal growth phenotypes. In the Tatton-Brown-Rahman syndrome, loss-of-function mutations in DNMT3A cause an overgrowth condition paired with mild to moderate intellectual disability [78] (table 1). Interestingly, these mutations are distributed over the PWWP, ADD, and MTase domains, but do not appear in the N-terminal stretch or the regions in between the domains. This suggests that they could interfere with the binding to histone modifications, domain-domain interactions, or both [78]. Reciprocally, recent studies linked gain-of-function mutations in the PWWP domain of DNMT3A to growth retardations [79, 80] (table 1). Whole-exome sequencing of microcephalic dwarfism patients identified two different de novo mutations, affecting two amino acids (W330R and D333N) that are part of the aromatic cage that forms around the methylated lysine 36 on histone H3 [82]. Mouse models carrying the murine equivalents of these mutations (W326R and D329A, respectively) displayed growth deficits as demonstrated by reduced brain size and body weight, recapitulating the human disease phenotype [79, 80]. On the molecular level, W326R and D329A were shown in in vitro experiments to impair binding of the PWWP domain to H3K36me2/3, suggesting altered binding specificity of DNMT3A to the DNA [57, 79]. Indeed, genome-wide assessment of DNA methylation in two microcephaly patient-derived fibroblast lines identified 1878 common differentially methylated regions (DMRs), the majority of which was hypermethylated relative to healthy controls [79]. Such hypermethylated DMRs were frequently positioned close to key developmental gene loci that were associated with Polycomb-mediated repression in control cells, but showed reduced levels of H3K27me3 in patient fibroblasts [79]. Similar results were obtained by genome-wide DNA methylation profiling in several tissues of adult Dnmt3aΔ/D329A mice [80]. Counterintuitively, increased DNA methylation at these sites leads to de-repression of the associated genes, possibly through displacement of the repressive Polycomb complexes [80]. Interestingly, gain-of-function mutations in the PWWP domain of DNMT3A have also been found in patients with paraganglioma, a type of neuroendocrine tumours [81] (table 1). CRISPR/Cas9-mediated introduction of one of these mutations (K299I) in HeLa cells caused genome-wide alterations in the DNA methylation landscape, suggesting that aberrant DNMT3A recruitment through mutations in the PWWP domain may be a more general disease-causing event [81].
Although DNMT3B is much less implicated in human disease, mutations in the DNMT3B gene are well described to cause ICF syndrome, a rare autosomal recessive disease that is characterised by immunodeficiency, centromeric instability, and facial anomalies [83] (table 1). These mutations have been found not only in the MTase domain, but also in the PWWP domain; one of which (S282P in human, S277P in mouse) was shown by ChIP-seq to abrogate interaction with H3K36me3 [59, 84]. A crystallography study subsequently revealed that this residue stabilises the interaction between methylated H3K36 and a conserved water molecule, building a strong hydrogen bond that is needed for DNMT3B-PWWP substrate recognition [82]. This indicates that H3K36me3-mediated DNMT3B recruitment is essential for healthy cell function.
Recruitment of DNMTs to their genomic target sites is a multifaceted process that is dependent on various epigenetic events, which both directly and indirectly contribute to the establishment and maintenance of the DNA methylation landscape. Many such contributing factors have been described, providing important insights into the underlying mechanisms, but at the same time leaving many questions unanswered. Indeed, further research is needed to address these outstanding issues and help construct a complete picture.
For DNMT1, the main histone modifications influencing its recruitment are H3R2, H3Ub, and H3K9me2/3. Not much is known about the gene-regulatory effect of H3R2, methylation of which has been reported to be either associated with euchromatin, or with heterochromatin through antagonising H3K4me3 [85, 86]. The role of H3Ub in transcriptional regulation is likewise not well characterised. In contrast, H3K9 methylation has been studied in more detail, revealing a link to repressed chromatin such as centromeres, transposons, repetitive elements, and imprinted loci [87]. H3K9me2/3 recognition by UHRF1 and subsequent DNMT1 recruitment or by DNMT1 itself thus provides a clear connection between this histone mark and DNA methylation at heterochromatic regions, whose cooperative effect is gene silencing.
The most important players influencing DNMT3 recruitment are H3K4 methylation, H3K27me3, and H3K36me2/3 (fig. 2). H3K4 methylation is a marker for regulatory regions, with H3K4me1 designating enhancers and H3K4me3 promoters [88]. As such, H3K4me3 is enriched at CGIs and anticorrelates with DNA methylation [87]. Interestingly, a subset of CGI promoters is decorated with the activating H3K4me3 and the repressive H3K27me3 histone marks at the same time. This combination has been termed ‘bivalent’, and is believed to ready key developmental genes for rapid activation or repression in a lineage-specific manner [87]. The presence of H3K27me3 by itself is associated with silencing and high levels of CpG methylation throughout most of the genome [62, 88]. At CGIs, however, H3K27me3 and DNA methylation are mutually exclusive, providing an explanation for the unmethylated status of bivalent promoters. Upon global reduction of DNA methylation, H3K27me3 peaks at these sites flatten and spread out, leading to an altered 3D genome through the loss of Polycomb-mediated distal interactions and suggesting that DNA methylation serves to restrict H3K27me3 domains at CGIs [62, 89–91]. The molecular mechanisms behind this redistribution of H3K27me3 in the absence of DNA methylation are hitherto unknown, and also the biological consequences remain an active area of investigation. Additionally, much remains unclear about the role of H3K27me3 in recruiting the DNMT3s, and DNMT3A in particular. To our current knowledge, no domain or region responsible for H3K27me3 recognition has been identified in any of the DNMT3 proteins, arguing against a direct interaction through substrate binding. The fact that the co-occurrence of H3K27me3 and DNA methylation is context-dependent likewise points to more indirect pathways. Future research should therefore investigate which players could constitute the missing link between H3K27me3 and DNMT recruitment.
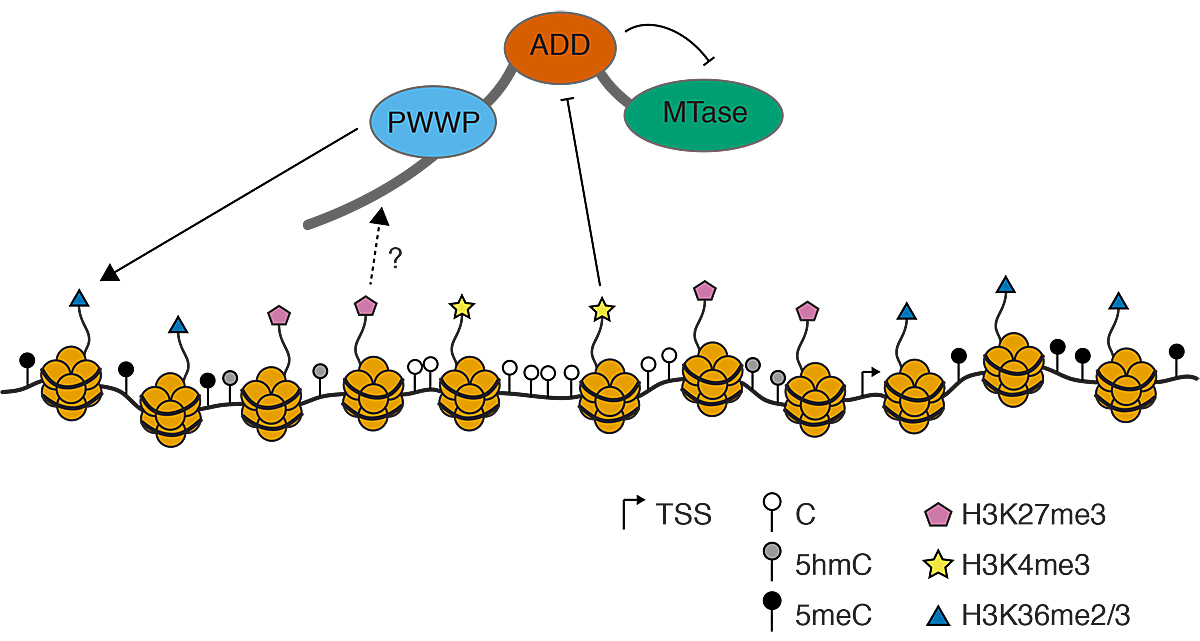
Figure 2 Histone tail modifications influencing DNMT3A/B recruitment to DNA. The PWWP domain recognises H3K36me2/3 (blue triangles), while the ADD domain is repelled by H3K4me3 (yellow stars). Additionally, the ADD domain auto-inhibits the MTase domain. How H3K27me3 (pink pentagon) influences DNMT3A/B recruitment remains to be elucidated. TSS = transcription start site
While H3K4me3 and H3K27me3 are important regulators for DNMT3 localisation at CGIs, H3K36me2/3 is strongly enriched across the transcribed bodies of active genes [87]. Both H3K36me2 and H3K36me3 positively correlate with DNA methylation, supporting their function in recruiting DNMT3A and DNMT3B, respectively [58, 59]. Recent studies described mutations in the PWWP domain of DNMT3A that abrogate H3K36 binding, instead recruiting this protein to chromatin that is marked with H3K27me3 in wildtype cells [79, 80]. Previous research had reported that such lowly methylated H3K27me3-marked regions are more likely to contract in the absence of DNMT3A, strengthening the notion that there is crosstalk between DNA methylation and H3K27me3 [92]. Whether this recruitment of mutated DNMT3A is specific or rather a general consequence of increased protein availability is still an open question, the answer to which will teach us more about the dynamics between H3K36me2/3 and H3K27me3 in instructing DNA methylation patterns.
Although recent research has significantly improved our structural understanding of the above-mentioned molecular mechanisms underlying DNMT recruitment, some important information is still missing. Since all structures of DNMT1 that are currently available have been obtained using truncated proteins, the exact domain arrangement of the full-length protein may not have been properly recapitulated [93]. This is especially relevant since interactions between different domains exist that influence the function of the protein, most notably the auto-inhibitory binding of the RFTS domain to the MTase domain [20]. For DNMT3A and DNMT3B it would likewise be useful to compare different stretches of the proteins, which would provide information not only on the function of the domains by themselves or in combination, but also on the naturally occurring isoforms. Additionally, advances in Cryo-EM could help to obtain novel mechanistic insights into DNMT function in the presence of their natural binding substrates of reconstituted nucleosomes carrying distinct modifications.
Furthermore, it would be informative to compare structures of the DNMTs interacting with other family members. Apart from interactions within the DNMT3 branch, DNMT3s are also known to interact with DNMT1 [94], yet if and how these interactions influence conformation and subsequent DNA binding as well as biological function remains to be determined. Such insights could in turn also shed light on the observation that different DNMT3 family members, but also protein isoforms, show differential DNA binding patterns [59, 60]. Besides recruitment of DNMTs to specific chromatin states, recent work suggested structural differences in substrate recognition that could contribute to differential DNMT3A/B activities based on flanking-DNA-sequence around the methylated cytosine [47–49]. Such DNA sequence-mediated differences could strongly contribute to global DNA methylation patterns and provide an additional layer of DNMT regulation, which should be further explored. Some hypotheses on this topic can nonetheless already be formulated based on our current knowledge, for example concerning the DNMT3A/B-DNMT3L interaction. As described, DNMT3L contributes to H3K4me0 binding through its ADD domain, but because of its lacking PWWP domain by definition cannot recognise H3K36 methylation [50]. We can therefore hypothesise that interaction with DNMT3L could strengthen the affinity of DNMT3A or DNMT3B for H3K4me0, while methylation of H3K36 may be less important. It would be interesting to investigate whether the methylation status of H3K4 is indeed more instructive for DNA methylation patterns than that of H3K36, and if this is correlated to the expression of DNMT3L.
All in all, although various pathways influencing DNMT recruitment to DNA have been charted, how exactly they should be navigated remains to be determined. Dissecting these mechanisms will aid our understanding of the crosstalk between different histone modifications and DNA methylation, and how this together influences the genome-wide epigenetic landscape. Yet this is still only part of the whole story, since factors like protein-protein interactions, post-translational modifications, and RNAs have also been described to be involved in recruiting DNMTs to their genomic target loci [93]. In addition to mechanisms that recruit DNMTs to the genome, allosteric regulation [93] and structural constrains that influence catalytic activity based on DNA sequence context [47, 95, 96] are expected to contribute to DNA methylation regulation. Continued efforts will be directed towards solving this intriguing puzzle.
We would like to apologise to all colleagues whose work could not be cited due to space limitations. Research in the laboratory of TB is supported by Swiss National Science Foundation Professorships, Sinergia and SPARK programmes; the University of Zurich and the EMBO Young Investigator Programme.
No financial support and no other potential conflict of interest relevant to this article was reported.
1 Culp LA , Dore E , Brown GM . Methylated bases in DNA of animal origin. Arch Biochem Biophys. 1970;136(1):73–9. doi:.https://doi.org/10.1016/0003-9861(70)90328-0
2 Lister R , Pelizzola M , Dowen RH , Hawkins RD , Hon G , Tonti-Filippini J , et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. doi:.https://doi.org/10.1038/nature08514
3 Bird A . The dinucleotide CG as a genomic signalling module. J Mol Biol. 2011;409(1):47–53. doi:.https://doi.org/10.1016/j.jmb.2011.01.056
4 Bird AP . CpG-rich islands and the function of DNA methylation. Nature. 1986;321(6067):209–13. doi:.https://doi.org/10.1038/321209a0
5 Mohandas T , Sparkes RS , Shapiro LJ . Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211(4480):393–6. doi:.https://doi.org/10.1126/science.6164095
6 Li E , Beard C , Jaenisch R . Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–5. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8247133. doi:.https://doi.org/10.1038/366362a0
7 Borgel J , Guibert S , Li Y , Chiba H , Schübeler D , Sasaki H , et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42(12):1093–100. doi:.https://doi.org/10.1038/ng.708
8 Walsh CP , Chaillet JR , Bestor TH . Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20(2):116–7. doi:.https://doi.org/10.1038/2413
9 Barau J , Teissandier A , Zamudio N , Roy S , Nalesso V , Hérault Y , et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016;354(6314):909–12. doi:.https://doi.org/10.1126/science.aah5143
10 Hata K , Okano M , Lei H , Li E . Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–93.
11 Goll MG , Bestor TH . Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74(1):481–514. doi:.https://doi.org/10.1146/annurev.biochem.74.010904.153721
12 Kriaucionis S , Heintz N . The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–30. doi:.https://doi.org/10.1126/science.1169786
13 Tahiliani M , Koh KP , Shen Y , Pastor WA , Bandukwala H , Brudno Y , et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. doi:.https://doi.org/10.1126/science.1170116
14 Ginno PA , Gaidatzis D , Feldmann A , Hoerner L , Imanci D , Burger L , et al. A genome-scale map of DNA methylation turnover identifies site-specific dependencies of DNMT and TET activity. Nat Commun. 2020;11(1):2680. doi:.https://doi.org/10.1038/s41467-020-16354-x
15 Charlton J , Jung EJ , Mattei AL , Bailly N , Liao J , Martin EJ , et al. TETs compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers. Nat Genet. 2020;52(8):819–27. doi:.https://doi.org/10.1038/s41588-020-0639-9
16 Ambrosi C , Manzo M , Baubec T . Dynamics and Context-Dependent Roles of DNA Methylation. J Mol Biol. 2017;429(10):1459–75. doi:.https://doi.org/10.1016/j.jmb.2017.02.008
17 Kouzarides T . Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi:.https://doi.org/10.1016/j.cell.2007.02.005
18 Cedar H , Bergman Y . Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi:.https://doi.org/10.1038/nrg2540
19 Bestor T , Laudano A , Mattaliano R , Ingram V . Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203(4):971–83. doi:.https://doi.org/10.1016/0022-2836(88)90122-2
20 Takeshita K , Suetake I , Yamashita E , Suga M , Narita H , Nakagawa A , et al. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc Natl Acad Sci USA. 2011;108(22):9055–9. doi:.https://doi.org/10.1073/pnas.1019629108
21 Rountree MR , Bachman KE , Baylin SB . DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25(3):269–77. doi:.https://doi.org/10.1038/77023
22 Gruenbaum Y , Cedar H , Razin A . Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295(5850):620–2. doi:.https://doi.org/10.1038/295620a0
23 Yoder JA , Soman NS , Verdine GL , Bestor TH . DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270(3):385–95. doi:.https://doi.org/10.1006/jmbi.1997.1125
24 Arand J , Spieler D , Karius T , Branco MR , Meilinger D , Meissner A , et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8(6):e1002750. doi:.https://doi.org/10.1371/journal.pgen.1002750
25 Dodge JE , Okano M , Dick F , Tsujimoto N , Chen T , Wang S , et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280(18):17986–91. doi:.https://doi.org/10.1074/jbc.M413246200
26 Feng J , Zhou Y , Campbell SL , Le T , Li E , Sweatt JD , et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13(4):423–30. doi:.https://doi.org/10.1038/nn.2514
27 Jeltsch A , Jurkowska RZ . New concepts in DNA methylation. Trends Biochem Sci. 2014;39(7):310–8. doi:.https://doi.org/10.1016/j.tibs.2014.05.002
28 Dahlet T , Argüeso Lleida A , Al Adhami H , Dumas M , Bender A , Ngondo RP , et al. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat Commun. 2020;11(1):3153. doi:.https://doi.org/10.1038/s41467-020-16919-w
29 Chuang LSH , Ian HI , Koh TW , Ng HH , Xu G , Li BFL . Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277(5334):1996–2000. doi:.https://doi.org/10.1126/science.277.5334.1996
30 Spada F , Haemmer A , Kuch D , Rothbauer U , Schermelleh L , Kremmer E , et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176(5):565–71. doi:.https://doi.org/10.1083/jcb.200610062
31 Sharif J , Muto M , Takebayashi S , Suetake I , Iwamatsu A , Endo TA , et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–12. doi:.https://doi.org/10.1038/nature06397
32 Arita K , Ariyoshi M , Tochio H , Nakamura Y , Shirakawa M . Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455(7214):818–21. doi:.https://doi.org/10.1038/nature07249
33 Li T , Wang L , Du Y , Xie S , Yang X , Lian F , et al. Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 2018;46(6):3218–31. doi:.https://doi.org/10.1093/nar/gky104
34 Karagianni P , Amazit L , Qin J , Wong J . ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28(2):705–17. doi:.https://doi.org/10.1128/MCB.01598-07
35 Nady N , Lemak A , Walker JR , Avvakumov GV , Kareta MS , Achour M , et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem. 2011;286(27):24300–11. doi:.https://doi.org/10.1074/jbc.M111.234104
36 Rajakumara E , Wang Z , Ma H , Hu L , Chen H , Lin Y , et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43(2):275–84. doi:.https://doi.org/10.1016/j.molcel.2011.07.006
37 Ren W , Fan H , Grimm SA , Guo Y , Kim JJ , Li L , et al. Direct readout of heterochromatic H3K9me3 regulates DNMT1-mediated maintenance DNA methylation. bioRxiv. 2020;2020:04.27.064493. https://www.biorxiv.org/content/10.1101/2020.04.27.064493v1.full.
38 Ishiyama S , Nishiyama A , Saeki Y , Moritsugu K , Morimoto D , Yamaguchi L , et al. Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance. Mol Cell. 2017;68(2):350–360.e7. doi:.https://doi.org/10.1016/j.molcel.2017.09.037
39 Nishiyama A , Mulholland CB , Bultmann S , Kori S , Endo A , Saeki Y , et al. Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat Commun. 2020;11(1):1222. doi:.https://doi.org/10.1038/s41467-020-15006-4
40 Molaro A , Malik HS , Bourc’his D . Dynamic evolution of de novo DNA methyltransferases in rodent and primate genomes. Mol Biol Evol. 2020;37(7):1882–92. doi:.https://doi.org/10.1093/molbev/msaa044
41 Aapola U , Shibuya K , Scott HS , Ollila J , Vihinen M , Heino M , et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65(3):293–8. doi:.https://doi.org/10.1006/geno.2000.6168
42 Bourc’his D , Xu GL , Lin CS , Bollman B , Bestor TH . Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294(5551):2536–9. doi:.https://doi.org/10.1126/science.1065848
43 Suetake I , Shinozaki F , Miyagawa J , Takeshima H , Tajima S . DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279(26):27816–23. doi:.https://doi.org/10.1074/jbc.M400181200
44 Okano M , Bell DW , Haber DA , Li E . DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi:.https://doi.org/10.1016/S0092-8674(00)81656-6
45 Okano M , Xie S , Li E . Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–20. doi:.https://doi.org/10.1038/890
46 Zhang ZM , Lu R , Wang P , Yu Y , Chen D , Gao L , et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018;554(7692):387–91. doi:.https://doi.org/10.1038/nature25477
47 Lin C-C , Chen Y-P , Yang W-Z , Shen JCK , Yuan HS . Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B. Nucleic Acids Res. 2020;48(7):3949–61. doi:.https://doi.org/10.1093/nar/gkaa111
48 Gao L , Emperle M , Guo Y , Grimm SA , Ren W , Adam S , et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat Commun. 2020;11(1):3355. doi:.https://doi.org/10.1038/s41467-020-17109-4
49 Anteneh H , Fang J , Song J . Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation. Nat Commun. 2020;11(1):2294. doi:.https://doi.org/10.1038/s41467-020-16213-9
50 Ooi SKT , Qiu C , Bernstein E , Li K , Jia D , Yang Z , et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–7. doi:.https://doi.org/10.1038/nature05987
51 Otani J , Nankumo T , Arita K , Inamoto S , Ariyoshi M , Shirakawa M . Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10(11):1235–41. doi:.https://doi.org/10.1038/embor.2009.218
52 Zhang Y , Jurkowska R , Soeroes S , Rajavelu A , Dhayalan A , Bock I , et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38(13):4246–53. doi:.https://doi.org/10.1093/nar/gkq147
53 Noh KM , Wang H , Kim HR , Wenderski W , Fang F , Li CH , et al. Engineering of a Histone-Recognition Domain in Dnmt3a Alters the Epigenetic Landscape and Phenotypic Features of Mouse ESCs. Mol Cell. 2015;59(1):89–103. doi:.https://doi.org/10.1016/j.molcel.2015.05.017
54 Okitsu CY , Hsieh C-L . DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27(7):2746–57. doi:.https://doi.org/10.1128/MCB.02291-06
55 Weber M , Hellmann I , Stadler MB , Ramos L , Pääbo S , Rebhan M , et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–66. doi:.https://doi.org/10.1038/ng1990
56 Guo X , Wang L , Li J , Ding Z , Xiao J , Yin X , et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517(7536):640–4. doi:.https://doi.org/10.1038/nature13899
57 Dhayalan A , Rajavelu A , Rathert P , Tamas R , Jurkowska RZ , Ragozin S , et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285(34):26114–20. doi:.https://doi.org/10.1074/jbc.M109.089433
58 Weinberg DN , Papillon-Cavanagh S , Chen H , Yue Y , Chen X , Rajagopalan KN , et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573(7773):281–6. doi:.https://doi.org/10.1038/s41586-019-1534-3
59 Baubec T , Colombo DF , Wirbelauer C , Schmidt J , Burger L , Krebs AR , et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520(7546):243–7. doi:.https://doi.org/10.1038/nature14176
60 Manzo M , Wirz J , Ambrosi C , Villaseñor R , Roschitzki B , Baubec T . Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J. 2017;36(23):3421–34. doi:.https://doi.org/10.15252/embj.201797038
61 Gu T , Lin X , Cullen SM , Luo M , Jeong M , Estecio M , et al. DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol. 2018;19(1):88. doi:.https://doi.org/10.1186/s13059-018-1464-7
62 Brinkman AB , Gu H , Bartels SJJ , Zhang Y , Matarese F , Simmer F , et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22(6):1128–38. doi:.https://doi.org/10.1101/gr.133728.111
63 Epsztejn-Litman S , Feldman N , Abu-Remaileh M , Shufaro Y , Gerson A , Ueda J , et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15(11):1176–83. doi:.https://doi.org/10.1038/nsmb.1476
64 Viré E , Brenner C , Deplus R , Blanchon L , Fraga M , Didelot C , et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–4. doi:.https://doi.org/10.1038/nature04431
65 Neri F , Krepelova A , Incarnato D , Maldotti M , Parlato C , Galvagni F , et al. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155(1):121–34. doi:.https://doi.org/10.1016/j.cell.2013.08.056
66 Rush M , Appanah R , Lee S , Lam LL , Goyal P , Lorincz MC . Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics. 2009;4(6):404–14. doi:.https://doi.org/10.4161/epi.4.6.9392
67 Hagarman JA , Motley MP , Kristjansdottir K , Soloway PD . Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLoS One. 2013;8(1):e53880. doi:.https://doi.org/10.1371/journal.pone.0053880
68 Veland N , Lu Y , Hardikar S , Gaddis S , Zeng Y , Liu B , et al. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 2019;47(1):152–67. doi:.https://doi.org/10.1093/nar/gky947
69 Fuks F , Hurd PJ , Deplus R , Kouzarides T . The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31(9):2305–12. doi:.https://doi.org/10.1093/nar/gkg332
70 Li H , Rauch T , Chen ZX , Szabó PE , Riggs AD , Pfeifer GP . The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281(28):19489–500. doi:.https://doi.org/10.1074/jbc.M513249200
71 Li E , Bestor TH , Jaenisch R . Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–26. doi:.https://doi.org/10.1016/0092-8674(92)90611-F
72 Klein CJ , Botuyan MV , Wu Y , Ward CJ , Nicholson GA , Hammans S , et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43(6):595–600. doi:.https://doi.org/10.1038/ng.830
73 Winkelmann J , Lin L , Schormair B , Kornum BR , Faraco J , Plazzi G , et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21(10):2205–10. doi:.https://doi.org/10.1093/hmg/dds035
74 Yang L , Rau R , Goodell MA . DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15(3):152–65. doi:.https://doi.org/10.1038/nrc3895
75 Roller A , Grossmann V , Bacher U , Poetzinger F , Weissmann S , Nadarajah N , et al. Landmark analysis of DNMT3A mutations in hematological malignancies. Leukemia. 2013;27(7):1573–8. doi:.https://doi.org/10.1038/leu.2013.65
76 Buscarlet M , Provost S , Zada YF , Barhdadi A , Bourgoin V , Lépine G , et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130(6):753–62. doi:.https://doi.org/10.1182/blood-2017-04-777029
77 Jaiswal S , Libby P . Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol. 2020;17(3):137–44. doi:.https://doi.org/10.1038/s41569-019-0247-5
78 Tatton-Brown K , Seal S , Ruark E , Harmer J , Ramsay E , Del Vecchio Duarte S , et al.; Childhood Overgrowth Consortium. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46(4):385–8. doi:.https://doi.org/10.1038/ng.2917
79 Heyn P , Logan CV , Fluteau A , Challis RC , Auchynnikava T , Martin CA , et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat Genet. 2019;51(1):96–105. doi:.https://doi.org/10.1038/s41588-018-0274-x
80 Sendžikaitė G , Hanna CW , Stewart-Morgan KR , Ivanova E , Kelsey G . A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat Commun. 2019;10(1):1884. doi:.https://doi.org/10.1038/s41467-019-09713-w
81 Remacha L , Currás-Freixes M , Torres-Ruiz R , Schiavi F , Torres-Pérez R , Calsina B , et al. Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet Med. 2018;20(12):1644–51. doi:.https://doi.org/10.1038/s41436-018-0003-y
82 Rondelet G , Dal Maso T , Willems L , Wouters J . Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J Struct Biol. 2016;194(3):357–67. doi:.https://doi.org/10.1016/j.jsb.2016.03.013
83 Xu GL , Bestor TH , Bourc’his D , Hsieh CL , Tommerup N , Bugge M , et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402(6758):187–91. doi:.https://doi.org/10.1038/46052
84 Shirohzu H , Kubota T , Kumazawa A , Sado T , Chijiwa T , Inagaki K , et al. Three novel DNMT3B mutations in Japanese patients with ICF syndrome. Am J Med Genet. 2002;112(1):31–7. doi:.https://doi.org/10.1002/ajmg.10658
85 Kirmizis A , Santos-Rosa H , Penkett CJ , Singer MA , Vermeulen M , Mann M , et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449(7164):928–32. doi:.https://doi.org/10.1038/nature06160
86 Migliori V , Müller J , Phalke S , Low D , Bezzi M , Mok WC , et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19(2):136–44. doi:.https://doi.org/10.1038/nsmb.2209
87 Mikkelsen TS , Ku M , Jaffe DB , Issac B , Lieberman E , Giannoukos G , et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi:.https://doi.org/10.1038/nature06008
88 Barski A , Cuddapah S , Cui K , Roh TY , Schones DE , Wang Z , et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi:.https://doi.org/10.1016/j.cell.2007.05.009
89 Reddington JP , Perricone SM , Nestor CE , Reichmann J , Youngson NA , Suzuki M , et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14(3):R25. doi:.https://doi.org/10.1186/gb-2013-14-3-r25
90 Joshi O , Wang SY , Kuznetsova T , Atlasi Y , Peng T , Fabre PJ , et al. Dynamic Reorganization of Extremely Long-Range Promoter-Promoter Interactions between Two States of Pluripotency. Cell Stem Cell. 2015;17(6):748–57. doi:.https://doi.org/10.1016/j.stem.2015.11.010
91 McLaughlin K , Flyamer IM , Thomson JP , Mjoseng HK , Shukla R , Williamson I , et al. DNA methylation directs polycomb-dependent 3D genome re- organization in naive pluripotency. Cell Rep. 2019;29(7):1974–1985.e6. doi:.https://doi.org/10.1016/j.celrep.2019.10.031
92 Jeong M , Sun D , Luo M , Huang Y , Challen GA , Rodriguez B , et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet. 2014;46(1):17–23. doi:.https://doi.org/10.1038/ng.2836
93 Jeltsch A , Jurkowska RZ . Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucleic Acids Res. 2016;44(18):8556–75. doi:.https://doi.org/10.1093/nar/gkw723
94 Kim GD , Ni J , Kelesoglu N , Roberts RJ , Pradhan S . Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21(15):4183–95. doi:.https://doi.org/10.1093/emboj/cdf401
95 Handa V , Jeltsch A . Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J Mol Biol. 2005;348(5):1103–12. doi:.https://doi.org/10.1016/j.jmb.2005.02.044
96 Wienholz BL , Kareta MS , Moarefi AH , Gordon CA , Ginno PA , Chédin F . DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010;6(9):e1001106. doi:.https://doi.org/10.1371/journal.pgen.1001106
No financial support and no other potential conflict of interest relevant to this article was reported.