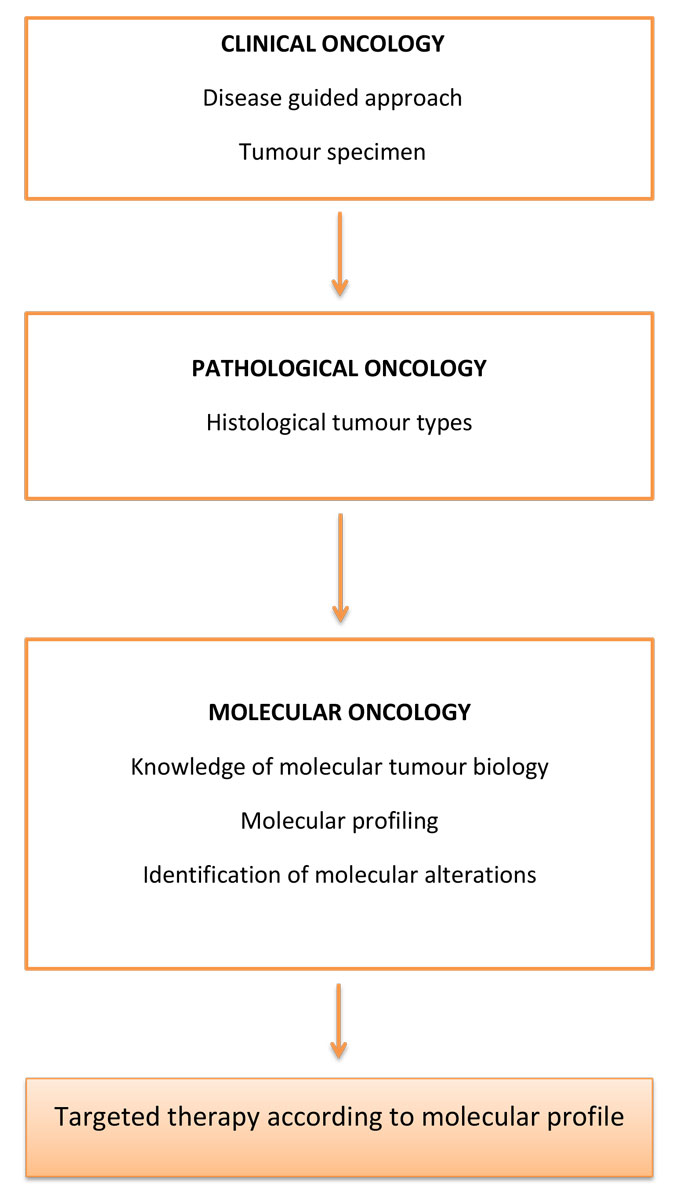
Figure 1 Conceptual evolution of oncological clinical practice.
DOI: https://doi.org/10.4414/smw.2019.20057
Despite the huge progress and efforts in cancer research to date, the war on cancer remains a major challenge and a never-ending battle. The global burden of cancer will increase to 22 million new cases each year by 2030 [1, 2]. One of the major obstacles to additional progress in the struggle against cancer is the complexity of the mechanisms underlying the disease. Unravelling cancer’s complexity by harnessing the power of cutting-edge technologies and computing tools has become a major objective of cancer research [3]. To this end, whole-genome sequencing and new measurement technologies let us pinpoint new therapeutic attack strategies [4]. The detection of alterations in gene-based structure and function could have major consequences for the diagnosis, prognosis and treatment of cancer [5]. Genome-analysis techniques provide data about genetic abnormalities, epigenetic changes and transcriptome modifications. Such work has led to a more integrated and inclusive approach to cancers and the accurate molecular characterisation of tumours [6]. Indeed, while cancers have long been classified according to clinical and histological signs, gene expression profiling leads to a new, genetic taxonomy of cancers according to their different molecular signatures. Such analyses are supposed to untangle the complex nature of cancer biology and to overcome the wide diversity of tumours through the identification of genes and pathways that could be deregulated to better detect and manage the disease [7]. Much effort is currently focused on the identification of molecular subtypes. These allow the best treatment option to be chosen, the risk of relapse to be predicted and therapeutic response to be assessed through a new, molecular taxonomy of cancer [8].
Major improvements in biotechnology and bioinformatics have been achieved over the past two decades, allowing the development of omic signatures of several molecular subtypes of cancer, each associated with different outcomes [9]. For instance, in breast cancer, which remains a devastating disease, with 5-year survival rates of metastatic breast cancer below 25%, several gene signatures have been devised which predict the risk of relapse, the response to specific treatments, and the benefit from adjuvant chemotherapy [10] [11]. As well as shedding light on cancer heterogeneity and enabling adequate treatment, genomic-profiling may lead to a better understanding of the metastatic process and foster other strategies for new drug development [12]. Therefore, several programs combining informatics, biomedical and biotechnology data aspire to create new ways to diagnose and treat cancers. The assessment of novel bioinformatic methodologies in support of genomics approaches may be the beginning of a new era in oncological clinical practice [13].
This review discusses the development of genomics approaches in cancer research and the potential of genomics for precision medicine, as well as clinical implications and remaining challenges.
The Cancer Genome Atlas (TCGA) project was launched in 2005 as an effort to accelerate our understanding of the molecular basis of cancer through the application of genome analysis technologies, including large-scale genome sequencing. The National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) jointly designed this major undertaking to improve the diagnosis, treatment and prevention of cancer through a complete identification of the molecular aberrations associated with cancer. The advent of polymerase chain reaction (PCR) and next-generation sequencing (NGS) have expanded the possibilities for genome analysis, methylation analysis and transcriptional profiling. Each biomolecular class, including RNA, DNA, proteins and lipids, has become the cornerstone of its own study, defining different omics approaches [14] [5]. The term genomics refers to DNA studies conducted through single-gene testing or multi-gene testing. The identification of all common cancer-related genomic aberrations would allow the assessment of tumours through their specific signatures. Genome sequencing data from TCGA project are now accessible for several tumours, including breast cancer, and should provide tools to assign the most appropriate therapeutic strategies in clinical practice [15]. Indeed, more than 11,000 patient samples and 33 tumour types are included in this database, with valuable data on key genomic changes providing a major contribution to the understanding of cancer genomics [16, 17]. Epigenetic features of cancer are the focus of another major project, the Roadmap Epigenomics Mapping Consortium, whose main objective is to generate a public human epigenomic database thanks to NGS technologies. The study of DNA methylation, histone modifications, RNA transcripts and chromatin accessibility aims to provide a reference of normal epigenomes that could help with the identification of relevant differences in comparison with epigenetics in cancer cells [18]. This work has led to the identification of the major genes and pathways involved in cancer genesis, as well as molecular events conferring oncogenic properties. It has also unveiled the full complexity of tumour heterogeneity between even histologically similar tumours [19]. Several other programs have since been formed, and the International Cancer Genome Consortium (ICGC) was created to coordinate a large number of research projects that have the common aim of comprehensively elucidating the genomic changes present in various forms of cancer. The main goal is to generate data for the entire research community on genomic abnormalities, including somatic mutations, abnormal expression of genes and epigenetic modifications, in tumours from different types of cancer. The ICGC data portal currently contains more than 24 cancer projects and copes with high computational volumes, high complexity and high heterogeneity of data [20]. The conceptual evolution of oncological clinical practice through high-throughput sequencing is described in figure 1 .

Figure 1 Conceptual evolution of oncological clinical practice.
Improvements in high-throughput DNA sequencing, along with computational and algorithmic advances, have led to tremendous progress in accessing omics data. NGS technologies provide accurate analyses of tumours’ pan-genomic profiles at genomic and transcriptomic levels, enabling the possibility of designing the right cancer drug for the right patient [21]. DNA sequencing technologies for human genomic medicine emerged more than 30 years ago and have undergone major technical improvements. Whereas the first automated genome sequencing machine (AB370), launched in 1987, was able to detect 96 bases simultaneously and 500,000 bases a day, the current AB3730 machine can detect up to 2.88 million bases per day [22, 23]. The first high-throughput NG sequencer, known as Illumina’s MiSeqDx, was launched in 2013 and paved the way for the development of new genome-based tests. Several NGS sequencing techniques, including whole genome sequencing, whole exome sequencing, transcriptome sequencing and targeted panel sequencing, have been designed since. NGS has become increasingly reliable, cheaper and faster, and allows the identification of crucial somatic mutations thanks to advances in nanotechnology and continued developments in bioinformatics [24]. Whole-genome sequencing sequences the complete genome of a sample, whereas whole-exome sequencing sequences the protein-coding genes. This approach aims to assess the full sequence of cancer-related gene panels. In another approach, certain regions of selected genes can be sequenced, focusing on cancer gene “hotspot” regions with recurrent mutations. The common goal remains the ability to perform almost any type of analysis in order to identify potential therapeutic targets at genomic and transcriptomic levels which can be used to classify tumours and predict outcomes [25]. Indeed, tumours are biologically diverse and contain complex genetic alterations. Well over 100 genes have been found which are frequently mutated in one kind of cancer or another, and the sheer number of cancer genes has frustrated attempts to deduce which ones are necessary and sufficient to cause the disease. Cancer genomes are highly rearranged, with several large-scale mutations including translocations, inversions, fusions and copy number changes [26]. In this respect, NGS could partly solve the partial map problem. Specific tools have been designed to process NGS data. Notably, several mapping software programs allow the analysis of tumour and normal genome pairs simultaneously, so that germline and somatic mutations can be distinguished [27] [28]. NGS data create huge bioinformatics challenges including storage, transmission, manipulation and analysis, making the downstream processing of these data a daunting task. DNA sequencing strategies are detailed in table 1 .
Table 1 Characteristics of DNA sequencing strategies.
| Whole genome sequencing | Whole exome sequencing | Targeted panels sequencing |
|---|---|---|
| Most comprehensive and unbiased examination of the cancer genome | Detection of 85% of disease-causing mutations | Rapid and reliable identification of the most common molecular aberrations |
| Discovery of new mutations | Detection of unknown variants High bioinformatics level |
Amplicon-based or hybridisation capture-based NGS |
| Most expensive | Higher depth of coverage Less expensive |
High depth of coverage and quick interpretation for clinical use |
| Uncertain value for clinical interpretation | Feasible in clinical routine and for research applications | Limited value for research and for complex aberrations |
While NGS allows lower cost, higher-throughput genome sequencing and opens up exciting new approaches in terms of personalised cancer therapy, the huge number and variety of genetic aberrations found in cancer creates substantial analytical complexity. However, while the abundance of information generated may complicate decisions regarding cancer therapy, genome sequencing has clearly led to the development of novel therapeutic targets through the identification of driver mutations [29]. The relevance and therapeutic consequences of specific mutations are assessed through biomarker-driven research that recruits selected patients into clinical trials to assess the efficacy of targeted therapies [30, 31]. Chen et al. have given an overview of the NGS work flow, from DNA isolation to the sequencing and data analysis which determine therapy. The ultimate goal is the identification of alterations to actionable genes which are associated with their functional impact and have therapeutic implications [32]. Glioblastoma, for instance, was the first cancer type to be sequenced and deposited into TCGA. The analysis of its transcriptome and genome signatures allowed the classification of these tumours into proneural, neural, classical and mesenchymal subtypes [33]. NGS clearly improves our ability to refine the nosology of tumours. Furthermore, mutations in chromatin-modifier genes, as well as in genes for which targeted therapies aimed at other diseases have been developed, including BRAF, FGFR1, FGFR2 and FGFR3, have been found in most glioblastoma tumours, demonstrating the potential clinical impact of such NGS data [34]. Potential targets for therapy were identified among a panel of 130 genes in more than 70% of glioblastomas [35]. Additionally, new drugs such as crizotinib have advanced to late-phase clinical trials for their anticancer effects on non-small cell lung cancers carrying EML4-ALK translocations [36]. Other therapies that target recurrent alterations, including EGFR mutations, MET amplification and ROS1 fusions, have also been developed in lung adenocarcinoma [37]. In fact, molecular profiling of lung cancer has become crucial for predicting the response to targeted therapies [38]. A landscape of driver mutations in melanoma has also been established, and BRAF V600 mutations, which are present in 50% of melanomas, predict clinical efficacy of RAF inhibitors such as vemurafenib, a tyrosine kinase inhibitor [39]. Furthermore, the ability of NGS to identify MEK mutations associated with resistance to vemurafenib may help to guide patient treatment. Similarly, the discovery of mutations in other driver oncogenes, such as NRAS, GNAQ and GNA11, could help to decide whether to use immunotherapy with CTLA4 or PDL1 inhibitors [38]. Analyses to identify drug targets and drug-resistance mutations are well documented for numerous cancer types. In a prospective study, Malapelle et al. recently identified mutations in KRAS, NRAS and BRAF which are associated with resistance to anti-EGFR therapy [40]. In head and neck cancer, distinct mutation profiles and targetable mutations could allow the identification of subgroups of patients with poor outcomes after adjuvant chemo radiation [41]. In triple negative breast cancer, integrated analysis of differentially expressed genes and pathways is ongoing. This aims to identify novel target molecules for therapy, as it remains a heterogeneous disease characterised by an aggressive phenotype and reduced survival [42]. A recent study using several NGS analyses on 439 patients with various types of cancer showed that 20% of patients had an actionable mutation targeted by on-label drugs, whereas 50% of patients had actionable mutations targeted by an off-label but approved drug [43]. Another prospective study of 800 patients demonstrated that more than 60% of patients had clinically relevant mutations, with 26% displaying a mutation with therapeutic implications [44]. Finally, a much more consequent study of 2,221 patients also concluded that relevant mutations were identified in 76% of cases [45–48]. These initial studies demonstrate the role of NGS in identifying a huge range of somatic alterations in cancer, leading to major therapeutic features.
Several clinical trials to assess the mutational profiles of cancer patients, especially metastatic ones, have been launched recently, leading to relevant results. In a Korean prospective trial which enrolled 407 patients between 2013 and 2014, 84% of patients had at least one aberration detected. When patients were matched to molecularly targeted agents, the response rate was significantly higher in the mutation-matched than in the non-matched treatment group [49]. The currently ongoing National Cancer Institute-Molecular Analysis for Therapy Choice (NCI-MATCH) trial is a national, signal-finding, precision medicine study that relies on genomic assays to screen and enrol patients with relapsed or refractory cancer after standard treatments using validated NGS processes [50]. The MPACT trial (molecular profiling-based assignment of cancer therapy; NCT01827384) was one of the first randomised studies to assess the response of patients treated with a drug matched to the mutational profile obtained through NGS analysis [51]. These underlying premises are clearly opening up a new era of genomic medicine. Besides the main goal of novel therapeutic targets specifically adapted to the features of a tumour, another important objective of omics technologies is to re-examine adjuvant strategies, especially in breast cancer, by finding molecular signatures to identify patients who need close monitoring rather than aggressive chemotherapies. Indeed, some gene signatures which predict the risk of relapse, the risk of distant metastasis or the response to specific therapies have been identified. In patients with breast cancer and a lymph-node-negative status, the 21-gene Oncotype Dx signature, the 70-gene MammaPrint signature and the 76-gene Rotterdam signature currently identify patients at high risk of developing distant metastases within five years. They show comparable performance, despite few genes being shared between them. The oncotype recurrence score in particular provides data concerning the benefit of adjuvant chemotherapy [52–55]. Such screening strategies offer a more sensitive detection of metastasis-prone cancers than only the immunohistochemical tumour characteristics used to classify breast cancer subtypes until now [56]. Improvements in outcomes may come either from new treatment strategies or from the identification of aggressive tumours which require dense and sharp therapeutic treatment, before metastatic dissemination. Other additional applications of NGS are still under development and include the evaluation of circulating tumour cells or free-plasma DNA to detect early relapse or residual cancer [57]. Once genomic alterations have been identified, PRC assays could be used to detect circulating tumour cells or free plasma DNA harbouring the same alterations, allowing the assessment of disease status, drug responsiveness and relapse. Accurate monitoring of the mutations harboured by a tumour may also allow the screening of new alterations that occur under treatment pressure, and thereby give insights into the mechanisms of acquired resistance [58]. Omics approaches are thus useful and promising tools to override the obstacle of tumour heterogeneity [59]. Another application of NGS is to improve cancer diagnosis in cases where histological identification is challenging through high sensitivity and specificity analysis, leading to the determination of the origin of the tumour [60]. Beyond genomics, the development of pharmacogenomics will also aid study of the association between genetic variation and anti-cancer drug response, as well as the development of predictive tools to anticipate tolerance and efficacy of treatment strategies, opening a totally new vision for therapeutics [61]. The prospects and challenges of NGS are summarised in figure 2 .
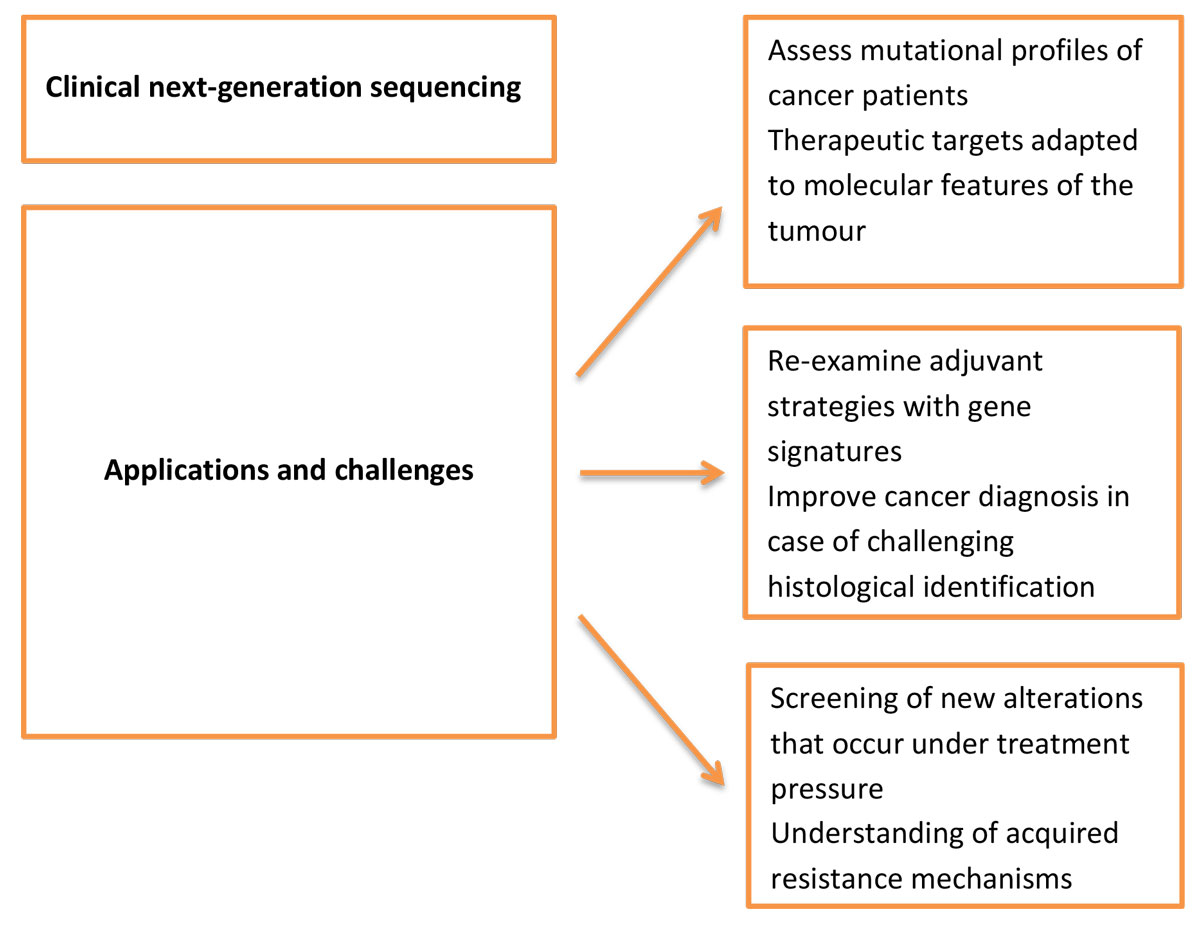
Figure 2 Prospective applications and challenges of next-generation sequencing.
Omics technologies have brought unprecedented advances in our understanding of the biology of cancer through the identification of relevant germline and somatic mutations. This has major clinical implications and paves the way to individualised medicine. Based on the massively parallel sequencing of DNA, with subsequent data processing and sequence alignment, NGS allows the simultaneous analysis of multiple genetic aberrations, including single nucleotide variants, small insertions/deletions, copy number variants and complex genomic rearrangements. The vast amounts of data generated by NGS have broadened our understanding of cancer. The concepts of omics, ranging from basic genomics to integrated systeomics, provide new insights into the genomics of tumour cells, which have subsequently led to crucial improvements in anti-cancer drugs [62]. In an article entitled “Cancer research: quo vadis – to war?”, Wheatley et al. emphasise that every cancer is as unique as the patient, and that each case must be seen as a challenge in itself. This is why there is such interest in omics technologies that open the way for a new era of genetic investigations which make cancer an idiosyncratic entity that should be treated as such [63].
The overall clinical potential of omics approaches in cancer research is already well known, but its utility probably goes beyond the hype, notably through the individualisation of treatment strategies. Long-term outcomes of better survival and clinically relevant benefits for patients are still being explored in clinical trials. Molecular databases are currently being updated to make it easier to understand the genetic profiles and to therefore provide specific and unique treatment recommendations. This novel paradigm will probably shape and enhance our understanding of tumour biology in the decades to come and be the cornerstone of the development of new anti-cancer drugs. The continued participation and collaboration of clinical oncologists, cancer researchers, computational biologists, bioinformaticians and, most importantly, patients remains the sine qua non condition for steady progress in genomic medicine. In default of the entire war, one part of the battle in the daunting fight against cancer may be about to be won [64].
No financial support and no other potential conflict of interest relevant to this article was reported.
1Bray F, Soerjomataram I. The Changing Global Burden of Cancer: Transitions in Human Development and Implications for Cancer Prevention and Control, The International Bank for Reconstruction and Development / The World Bank, 2015. doi:https://doi.org/10.1596/978-1-4648-0349-9_CH2
2 Sporn MB . The war on cancer. Lancet. 1996;347(9012):1377–81. doi:.https://doi.org/10.1016/S0140-6736(96)91015-6
3 Garland J . Unravelling the complexity of signalling networks in cancer: A review of the increasing role for computational modelling. Crit Rev Oncol Hematol. 2017;117:73–113. doi:.https://doi.org/10.1016/j.critrevonc.2017.06.004
4 Azizi E , Clouthier SG , Wicha MS . The Promise of Single Cell Omics for Onco-therapy. J Mol Genet Med. 2014;8(03). doi:.https://doi.org/10.4172/1747-0862.1000121
5 Epstein RJ , Lin FP . Cancer and the omics revolution. Aust Fam Physician. 2017;46(4):189–93.
6 Hinkson IV , Davidsen TM , Klemm JD , Kerlavage AR , Kibbe WA . A Comprehensive Infrastructure for Big Data in Cancer Research: Accelerating Cancer Research and Precision Medicine. Front Cell Dev Biol. 2017;5:83. doi:.https://doi.org/10.3389/fcell.2017.00083
7 Vucic EA , Thu KL , Robison K , Rybaczyk LA , Chari R , Alvarez CE , et al. Translating cancer ‘omics’ to improved outcomes. Genome Res. 2012;22(2):188–95. doi:.https://doi.org/10.1101/gr.124354.111
8 Grossman RL , Heath AP , Ferretti V , Varmus HE , Lowy DR , Kibbe WA , et al. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med. 2016;375(12):1109–12. doi:.https://doi.org/10.1056/NEJMp1607591
9 Griffith OL , Gray JW . 'Omic approaches to preventing or managing metastatic breast cancer. Breast Cancer Res. 2011;13(6):230. doi:.https://doi.org/10.1186/bcr2923
10 Siegel RL , Miller KD , Jemal A . Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi:.https://doi.org/10.3322/caac.21254
11 Coleman WB . Next-Generation Breast Cancer Omics. Am J Pathol. 2017;187(10):2130–2. doi:.https://doi.org/10.1016/j.ajpath.2017.07.011
12 Wood SL , Westbrook JA , Brown JE . Omic-profiling in breast cancer metastasis to bone: implications for mechanisms, biomarkers and treatment. Cancer Treat Rev. 2014;40(1):139–52. doi:.https://doi.org/10.1016/j.ctrv.2013.07.006
13 Yu K-H , Snyder M . Omics Profiling in Precision Oncology. Mol Cell Proteomics. 2016;15(8):2525–36. doi:.https://doi.org/10.1074/mcp.O116.059253
14 Gonzalez-Angulo AM , Hennessy BTJ , Mills GB . Future of personalized medicine in oncology: a systems biology approach. J Clin Oncol. 2010;28(16):2777–83. doi:.https://doi.org/10.1200/JCO.2009.27.0777
15 Kotelnikova EA , Pyatnitskiy M , Paleeva A , Kremenetskaya O , Vinogradov D . Practical aspects of NGS-based pathways analysis for personalized cancer science and medicine. Oncotarget. 2016;7(32):52493–516. doi:.https://doi.org/10.18632/oncotarget.9370
16Home - The Cancer Genome Atlas - Cancer Genome - TCGA. (n.d.). https://cancergenome.nih.gov/ (accessed February 14, 2018).
17 Giordano TJ . The cancer genome atlas research network: a sight to behold. Endocr Pathol. 2014;25(4):362–5. doi:.https://doi.org/10.1007/s12022-014-9345-4
18 Bernstein BE , Stamatoyannopoulos JA , Costello JF , Ren B , Milosavljevic A , Meissner A , et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–8. doi:.https://doi.org/10.1038/nbt1010-1045
19 Vucic EA , Thu KL , Robison K , Rybaczyk LA , Chari R , Alvarez CE , et al. Translating cancer ‘omics’ to improved outcomes. Genome Res. 2012;22(2):188–95. doi:.https://doi.org/10.1101/gr.124354.111
20 Zhang J , Baran J , Cros A , Guberman JM , Haider S , Hsu J , et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database (Oxford). 2011;2011(0):bar026–026. doi:.https://doi.org/10.1093/database/bar026
21 Buescher JM , Driggers EM . Integration of omics: more than the sum of its parts. Cancer Metab. 2016;4(1):4. doi:.https://doi.org/10.1186/s40170-016-0143-y
22 Bentley DR . Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16(6):545–52. doi:.https://doi.org/10.1016/j.gde.2006.10.009
23 Git A , Dvinge H , Salmon-Divon M , Osborne M , Kutter C , Hadfield J , et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16(5):991–1006. doi:.https://doi.org/10.1261/rna.1947110
24 Bahassi M , Stambrook PJ . Next-generation sequencing technologies: breaking the sound barrier of human genetics. Mutagenesis. 2014;29(5):303–10. doi:.https://doi.org/10.1093/mutage/geu031
25 Ulahannan D , Kovac MB , Mulholland PJ , Cazier J-B , Tomlinson I . Technical and implementation issues in using next-generation sequencing of cancers in clinical practice. Br J Cancer. 2013;109(4):827–35. doi:.https://doi.org/10.1038/bjc.2013.416
26 Chakravarthi BVSK , Nepal S , Varambally S . Genomic and Epigenomic Alterations in Cancer. Am J Pathol. 2016;186(7):1724–35. doi:.https://doi.org/10.1016/j.ajpath.2016.02.023
27 Lee H , Lee K-W , Lee T , Park D , Chung J , Lee C , et al. Performance evaluation method for read mapping tool in clinical panel sequencing. Genes Genomics. 2018;40(2):189–97. doi:.https://doi.org/10.1007/s13258-017-0621-9
28 Escalona M , Rocha S , Posada D . A comparison of tools for the simulation of genomic next-generation sequencing data. Nat Rev Genet. 2016;17(8):459–69. doi:. Correction published in: Nat Rev Genet. 2018;19:733. https://doi.org/10.1038/nrg.2016.57
29 Daber R , Sukhadia S , Morrissette JJD . Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet. 2013;206(12):441–8. doi:.https://doi.org/10.1016/j.cancergen.2013.11.005
30Molecular Oncology Tumor Boards | ASCO University, (n.d.). https://university.asco.org/motb (accessed February 21, 2018).
31 Harada S , Arend R , Dai Q , Levesque JA , Winokur TS , Guo R , et al. Implementation and utilization of the molecular tumor board to guide precision medicine. Oncotarget. 2017;8(34):57845–54. doi:.https://doi.org/10.18632/oncotarget.18471
32 Chen K , Meric-Bernstam F , Zhao H , Zhang Q , Ezzeddine N , Tang LY , et al. Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin Chem. 2015;61(3):544–53. doi:.https://doi.org/10.1373/clinchem.2014.231100
33 Brennan CW , Verhaak RGW , McKenna A , Campos B , Noushmehr H , Salama SR , et al.; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–77. doi:.. Erratum in: Cell. 2014;157(3):753. doi:https://doi.org/10.1016/j.cell.2013.09.034
34 Verhaak RGW , Hoadley KA , Purdom E , Wang V , Qi Y , Wilkerson MD , et al.; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi:.https://doi.org/10.1016/j.ccr.2009.12.020
35 Sahm F , Schrimpf D , Jones DTW , Meyer J , Kratz A , Reuss D , et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131(6):903–10. doi:.https://doi.org/10.1007/s00401-015-1519-8
36 Hallberg B , Palmer RH . Crizotinib--latest champion in the cancer wars? N Engl J Med. 2010;363(18):1760–2. doi:.https://doi.org/10.1056/NEJMe1010404
37 Camidge DR , Pao W , Sequist LV . Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11(8):473–81. doi:.https://doi.org/10.1038/nrclinonc.2014.104
38 Reiman A , Kikuchi H , Scocchia D , Smith P , Tsang YW , Snead D , et al. Validation of an NGS mutation detection panel for melanoma. BMC Cancer. 2017;17(1):150. doi:.https://doi.org/10.1186/s12885-017-3149-0
39 Hodis E , Watson IR , Kryukov GV , Arold ST , Imielinski M , Theurillat J-P , et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–63. doi:.https://doi.org/10.1016/j.cell.2012.06.024
40 Malapelle U , Vigliar E , Sgariglia R , Bellevicine C , Colarossi L , Vitale D , et al. Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. J Clin Pathol. 2015;68(1):64–8. doi:.https://doi.org/10.1136/jclinpath-2014-202691
41 Tinhofer I , Budach V , Saki M , Konschak R , Niehr F , Jöhrens K , et al.; DKTK-ROG. Targeted next-generation sequencing of locally advanced squamous cell carcinomas of the head and neck reveals druggable targets for improving adjuvant chemoradiation. Eur J Cancer. 2016;57:78–86. doi:.https://doi.org/10.1016/j.ejca.2016.01.003
42 Peng C , Ma W , Xia W , Zheng W . Integrated analysis of differentially expressed genes and pathways in triple‑negative breast cancer. Mol Med Rep. 2017;15(3):1087–94. doi:.https://doi.org/10.3892/mmr.2017.6101
43 Schwaederle M , Daniels GA , Piccioni DE , Fanta PT , Schwab RB , Shimabukuro KA , et al. On the Road to Precision Cancer Medicine: Analysis of Genomic Biomarker Actionability in 439 Patients. Mol Cancer Ther. 2015;14(6):1488–94. doi:.https://doi.org/10.1158/1535-7163.MCT-14-1061
44 Wong SQ , Fellowes A , Doig K , Ellul J , Bosma TJ , Irwin D , et al. Assessing the clinical value of targeted massively parallel sequencing in a longitudinal, prospective population-based study of cancer patients. Br J Cancer. 2015;112(8):1411–20. doi:.https://doi.org/10.1038/bjc.2015.80
45 Uzilov AV , Ding W , Fink MY , Antipin Y , Brohl AS , Davis C , et al. Development and clinical application of an integrative genomic approach to personalized cancer therapy. Genome Med. 2016;8(1):62. doi:.https://doi.org/10.1186/s13073-016-0313-0
46 Lane BR , Bissonnette J , Waldherr T , Ritz-Holland D , Chesla D , Cottingham SL , et al.; Spectrum Health Tumor Sequencing Advisory Board. Development of a Center for Personalized Cancer Care at a Regional Cancer Center: Feasibility Trial of an Institutional Tumor Sequencing Advisory Board. J Mol Diagn. 2015;17(6):695–704. doi:.https://doi.org/10.1016/j.jmoldx.2015.07.003
47 Boland GM , Piha-Paul SA , Subbiah V , Routbort M , Herbrich SM , Baggerly K , et al. Clinical next generation sequencing to identify actionable aberrations in a phase I program. Oncotarget. 2015;6(24):20099–110. doi:.https://doi.org/10.18632/oncotarget.4040
48 Tran B , Brown AMK , Bedard PL , Winquist E , Goss GD , Hotte SJ , et al. Feasibility of real time next generation sequencing of cancer genes linked to drug response: results from a clinical trial. Int J Cancer. 2013;132(7):1547–55. doi:.https://doi.org/10.1002/ijc.27817
49 Kim ST , Lee J , Hong M , Park K , Park JO , Ahn T , et al. The NEXT-1 (Next generation pErsonalized tX with mulTi-omics and preclinical model) trial: prospective molecular screening trial of metastatic solid cancer patients, a feasibility analysis. Oncotarget. 2015;6(32):33358–68. doi:.https://doi.org/10.18632/oncotarget.5188
50 Lih C-J , Harrington RD , Sims DJ , Harper KN , Bouk CH , Datta V , et al. Analytical Validation of the Next-Generation Sequencing Assay for a Nationwide Signal-Finding Clinical Trial. J Mol Diagn. 2017;19(2):313–27. doi:.https://doi.org/10.1016/j.jmoldx.2016.10.007
51 Lih C-J , Sims DJ , Harrington RD , Polley EC , Zhao Y , Mehaffey MG , et al. Analytical Validation and Application of a Targeted Next-Generation Sequencing Mutation-Detection Assay for Use in Treatment Assignment in the NCI-MPACT Trial. J Mol Diagn. 2016;18(1):51–67. doi:.https://doi.org/10.1016/j.jmoldx.2015.07.006
52 Foekens JA , Atkins D , Zhang Y , Sweep FCGJ , Harbeck N , Paradiso A , et al. Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol. 2006;24(11):1665–71. doi:.https://doi.org/10.1200/JCO.2005.03.9115
53 Wang Y , Klijn JG , Zhang Y , Sieuwerts AM , Look MP , Yang F , et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–9. doi:.https://doi.org/10.1016/S0140-6736(05)70933-8
54 van ’t Veer LJ , Dai H , van de Vijver MJ , He YD , Hart AAM , Mao M , et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi:.https://doi.org/10.1038/415530a
55 Paik S , Shak S , Tang G , Kim C , Baker J , Cronin M , et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi:.https://doi.org/10.1056/NEJMoa041588
56 Griffith OL , Gray JW . 'Omic approaches to preventing or managing metastatic breast cancer. Breast Cancer Res. 2011;13(6):230. doi:.https://doi.org/10.1186/bcr2923
57 Schwaederle M , Parker BA , Schwab RB , Fanta PT , Boles SG , Daniels GA , et al. Molecular tumor board: the University of California-San Diego Moores Cancer Center experience. Oncologist. 2014;19(6):631–6. doi:.https://doi.org/10.1634/theoncologist.2013-0405
58 Yap TA , Gerlinger M , Futreal PA , Pusztai L , Swanton C . Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4(127):127ps10. doi:.https://doi.org/10.1126/scitranslmed.3003854
59 Basho RK , Eterovic AK . Meric-Bernstam F . Clinical Applications and Limitations of Next-Generation Sequencing. Am J Hematol Oncol. 2015;11. Available at: https://www.gotoper.com/publications/ajho/2015/2015mar/clinical-applications-and-limitations-of-next-generation-sequencing
60 Nikiforov YE , Carty SE , Chiosea SI , Coyne C , Duvvuri U , Ferris RL , et al. Highly accurate diagnosis of cancer in thyroid nodules with follicular neoplasm/suspicious for a follicular neoplasm cytology by ThyroSeq v2 next-generation sequencing assay. Cancer. 2014;120(23):3627–34. doi:.https://doi.org/10.1002/cncr.29038
61 Yadav NK , Shukla P , Omer A , Pareek S , Srivastava AK , Bansode FW , et al. Erratum to “Next Generation Sequencing: Potential and Application in Drug Discovery”. Sci World J. 2014;2014:1. doi:.https://doi.org/10.1155/2014/621354
62Kulski JK. Next-Generation Sequencing — An Overview of the History, Tools, and “Omic” Applications, in: Next Gener. Seq. - Adv. Appl. Challenges, InTech, 2016. doi:https://doi.org/10.5772/61964.
63 Wheatley D . Cancer research: quo vadis-to war? Ecancermedicalscience. 2014;8:ed45. doi:.https://doi.org/10.3332/ecancer.2014.ed45
64 Horak P , Fröhling S , Glimm H . Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1(5):e000094. doi:.https://doi.org/10.1136/esmoopen-2016-000094
No financial support and no other potential conflict of interest relevant to this article was reported.