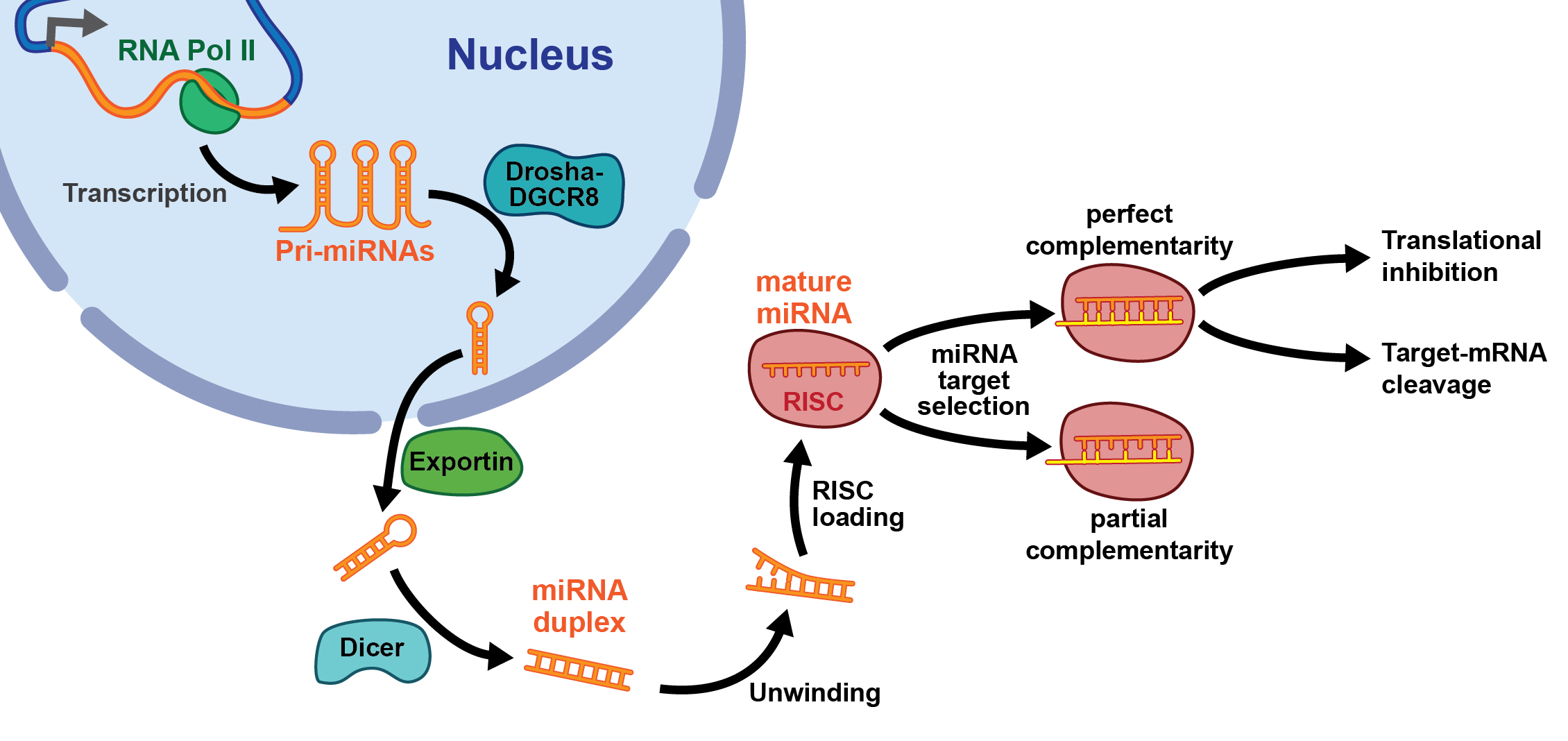
Figure 1 Biogenesis and function of microRNAs (see text). RISC = RNA-induced silencing complex
DOI: https://doi.org/10.4414/smw.2019.14703
Acute kidney injury (AKI) is associated with considerable morbidity and was identified previously as an independent risk factor for mortality of patients [1]. Even though significant advances have been made over the past several years regarding best supportive care, AKI on the intensive care unit is still highly prevalent and associated with a high mortality [2]. As evidenced by a major multinational, multicentre study involving more than 30,000 critically-ill patients with AKI on the intensive care unit, the in-hospital mortality exceeds 60% [3]. Thirteen percent of survivors who are subsequently discharged from hospital progress to end-stage kidney disease and remain on dialysis [3]. Ischaemia/reperfusion injury (I/R injury) of the kidney represents one of the major risk factors for the development of AKI [4]. A variety of different injurious insults in native kidneys (e.g., during cardiac surgery) culminate in I/R injury [4]. In addition, it is also an unavoidable phenomenon in transplanted kidneys owing to the transplantation procedure itself [5]. Because of its prevalence, it represents a major socioeconomic health problem [4]. During ischaemic AKI, a transient drop in blood flow to the kidney is followed by a reperfusion period. Reperfusion itself, though vital to restoration of kidney function, is associated with significant additional cellular injury [6]. Among a multitude of different signalling pathways, non-coding RNAs may also significantly contribute to the induction and resolution of renal I/R injury.
RNA transcripts without protein-coding potential represent more than 90% of the human genome [7, 8]. These noncoding RNA transcripts (ncRNAs) are arbitrarily separated into long ncRNAs (lncRNAs, ≥200 nucleotides) and small ncRNAs (≤200 nucleotides). The biogenesis and function of small ncRNAs such as microRNAs (miRNAs) has previously been described in detail [6–8]. In contrast, knowledge of the role and function of lncRNAs is largely lacking. The lncRNA class comprises both linear lncRNAs (named by default as lncRNAs) and circular RNAs (circRNAs). Current knowledge of the role of these three categories of ncRNA molecules (miRNAs, lncRNAs, circRNAs) in kidney function and ischaemic AKI is reviewed in the following paragraphs.
As mentioned in the introduction, during ischaemic AKI a transient drop in blood flow to the kidney is followed by a reperfusion period, which is associated with significant additional cellular injury [6]. The damage inflicted by tissue ischaemia is subsequently aggravated by a dramatic surge in reactive oxygen and nitrogen species during reperfusion [9, 10]. These induce protein modifications, lipid oxidation and DNA double strand breaks, finally culminating in endothelial dysfunction, neutrophil adherence to endothelium and transendothelial migration, the release of inflammatory mediators, cellular calcium overload and eventually cell death [9, 10]. In the kidney, blood flow to the outer medulla is disproportionately reduced compared with the reduction in total blood flow [9, 10]. Thus, epithelial cell injury is mainly detected in the S3 segment of the proximal tubule, located in the outer medulla [9, 10]. Interplay of several events contributes to the cellular injury observed in the kidney. The damaged endothelium interacts with and activates inflammatory cells through enhanced expression of adhesion molecules (e.g., intracellular adhesion molecule-1 [ICAM-1], selectins) [9, 10]. This interaction in turn contributes to obstruction of capillaries and postcapillary venules, further activation and transmigration of leucocytes, production of cytokines and inflammation in tubular epithelial cells [9, 10]. Capillary rarefaction in the inner stripe of the outer medulla ensues as a result of the development of chronic hypoxia and is an important contributor to post-AKI tubulointerstitial fibrosis and progression to chronic kidney disease [9, 10]. Cell polarity and cytoskeletal arrangement is severely impaired in proximal tubular epithelial cells during ischaemia [9, 10]. Important phenotypical changes are loss of the proximal tubule brush border as well as loss of polarity and derangement of adhesion molecules and other membrane proteins and disruption of cell-cell interactions at adherent and tight junctions [9, 10].
MicroRNAs (miRNAs) have been extensively studied over the past 10 years and found to have a major role in ischaemic AKI [11]. MiRNAs are small noncoding RNA transcripts with a length of approximately 22 nucleotides, which may be perceived as being responsible for fine tuning of protein expression by targeting the 3′-untranslated region of mRNA transcripts, thereby inducing transcriptional repression or transcript degradation [11]. The first miRNA, lin-4, was identified in Caenorhabditis elegans during investigation of genetic loci responsible for temporal control of postembryonic development [12, 13]. The number of identified mature miRNAs in humans exceeds 2000 (www.mirbase.org; release 21), thereby representing 1% of all human genes [14]. MiRNA biogenesis can be viewed as a two-step process (see fig. 1). Transcription of miRNA genes by RNA polymerase II results in primary miRNA transcripts, which are subsequently processed in the nucleus by the ribonuclease Drosha into precursor miRNAs with a length of 70 nucleotides [11]. Thereafter, a Ran-GTP-dependent nucleo/cytoplasmic cargo transporter named exportin 5 transports precursor miRNAs into the cytosol [11]. Here, they are further processed by a second ribonuclease, called Dicer, into a small, double-stranded RNA duplex, which is composed of an miRNA guide strand and its complementary passenger strand (miRNA*) [11]. In order to exert its function, the miRNA guide strand (in most cases) is incorporated into the RNA-induced silencing complex (RISC), where, with argonaute and other proteins, the 3′-untranslated region of mRNAs is targeted, thereby leading to repression of protein translation or degradation of mRNA [11]. In most cases, miRNA* is degraded, but rarely it can also function as a mature miRNA [11]. MiRNA function can be modulated by miRNA antagonists, so-called antimiRs (chemically engineered oligonucleotides targeting specific miRNAs) and miRNA mimics [11]. These miRNA modulators may have therapeutic potential in the treatment of patients with kidney disease, including AKI [11]. In addition to their intracellular regulation and role in the diagnosis and treatment of various diseases, they are also secreted into blood and urine and may therefore serve as biomarkers of disease and response to therapy [15].

Figure 1 Biogenesis and function of microRNAs (see text). RISC = RNA-induced silencing complex
In 2010, an initial study was published that investigated the role of miRNAs in AKI by modulation of the miRNA biogenesis machinery through specific deletion of Dicer in renal proximal tubular epithelium in mice. It was shown that after bilateral I/R injury of the kidney, Dicer knockout mice displayed preserved kidney function and reduced tissue damage compared with littermate controls [16]. MiR-192 was associated with AKI in rats [17]. We previously showed miR-24 to be enriched in the kidney following I/R injury in mice and humans [18]. MiR-24 was specifically enriched in renal endothelial and tubular epithelial cells after I/R induction. Through regulation of its targets H2A histone family, member X, the sphingosine-1-phopshate receptor 1, and haem oxygenase 1, miR-24 induced apoptosis of these cells. Silencing of miR-24 in mice following I/R injury resulted in a significant improvement of survival and kidney function, a reduction of apoptosis, improved histological tubular epithelial injury and less infiltration of inflammatory cells.
In unilateral renal I/R injury in mice, nine miRNAs were shown to be regulated differentially compared with control animals (miR-21, miR-20a, miR-146a, miR-199a-3p, miR-214, miR-192, miR-187, miR-805, and miR-194) [10, 19]. This signature was confirmed in immunodeficient recombination activating gene-2 (RAG-2) / common γ-chain double-knockout mice, suggesting that the miRNA expression is independent of influx of inflammatory cells [10, 19]. MiR-21 expression increased in proliferating tubular epithelial cells, and silencing of miR-21 promoted apoptosis [10, 20]. MiR-127 was demonstrated to be altered in a rat model of renal I/R injury. Here, miR-127 was postulated to be transcriptionally activated by hypoxia-inducible factor-1α (HIF-1α) and shown to target kinesin family member 3B (KIF3B) [10, 21]. Endothelial progenitor cells were shown to secrete miRNA-enriched microvesicles, which ameliorated murine renal I/R injury [10, 21]. Pro-angiogenic and anti-apoptotic miR-126 and miR-296 were found to be included in microvesicles. Injection of microvesicles derived from endothelial progenitor cells into rats subjected to renal I/R injury led to proliferation of tubular cells and a reduction in the number of apoptotic tubular cells, and decreased the infiltration of leucocytes [10, 22]. MiR-21 was found to contribute to renoprotection in a process termed ischaemic preconditioning [10, 23]. MiR-21 was associated with a significant amelioration, while antimir-21-mediated silencing promoted renal I/R injury [10, 23]. These effects were mediated by modulation of the miR-21 target programmed cell death protein 4 (PDCD4). MiR-21 was transcriptionally activated by HIF-1α [10, 23]. MiR-182-5p and miR-21-3p were associated with the development of AKI in kidney transplant patients [24]. In vivo silencing of miR-182-5p in a model of AKI in rats preserved renal function [25]. MiR-126 has previously been shown to have a beneficial effect regarding the development of AKI in mice [26]. Haematopoietic overexpression of miR-126 increased neovascularisation, preserved kidney function and increased numbers of bone marrow-derived endothelial cells. The numbers of circulating Lin−/Sca-1+/cKit+ haematopoietic stem and progenitor cells were increased [26].
Expression levels of miR-709, miR-217 and miR-696 have been shown to be altered in plasma and kidney tissue following bilateral renal I/R injury [27]. MiR-10a was elevated in plasma 1 hour after renal I/R injury in rats and thereby outperformed levels of creatinine (6 hours) and urea (12 hours) in detection of renal injury [28]. It was also previously shown to be altered in rat kidney following deep hypothermic circulatory arrest [29]. MiR-21 und -320 are increased in plasma and reduced in urine of rats following AKI [30]. The expression of miR-200a increases in the plasma and kidneys of mice following AKI [31]. We were able to show that miRNAs, specifically miR-210, detected in the plasma of critically ill patients with AKI serve as powerful and independent predictors of mortality in this patient cohort [32]. Urinary miRs were shown to be specifically decreased during acute T-cell mediated renal allograft rejection and to correlate with kidney function at 1 year after transplantation [33].
Relevant microRNAs in animals and human models of AKI are summarised in table 1.
Table 1 MicroRNAs, their targets and effects in renal ischaemia/reperfusion injury.
| MicroRNA | Target | Organ/cell type | Pathophysiological effects | Organism | Reference |
|---|---|---|---|---|---|
| miR-132, miR-362 and miR-379 | / | Kidney / proximal tubular cell | Dicer deletion | Mouse | [16] |
| miR-21, miR-20a, miR-146a, miR-199a-3p, miR-214, miR-192, miR-187, miR-805 and miR-194 | / | Kidney | Signature of kidney ischaemia/reperfusion injury | Mouse | [10, 19] |
| miR-127 | Kinesin family member 3B | Kidney / proximal tubular cell | Changes in cell adhesion and cytoskeleton structure | Rat | [21] |
| miR-126 and miR-296 | / | EPC-derived MVs / proximal tubular cell | Inhibition of capillary rarefaction, glomerulosclerosis, and tubulointerstitial fibrosis | Mouse | [22] |
| miR-21 | Programmed cell death protein 4 | Kidney / proximal tubular cell | Reduction of renal injury | Mouse | [23] |
| miR-192 | / | plasma | Biomarker of AKI | Rat | [17] |
| miR-24 | H2A.X, sphingosine 1-phosphate receptor 1, haem oxygenase-1 | Proximal tubular cell, endothelial cell | Amelioration of AKI | Mouse | [18] |
| miR-182-5p | / | / | Improvement of kidney function and morphology after AKI | Rat | [25] |
| miR-126 | / | Haematopoietic overexpression of miR-126 | increased neovascularisation, preserved kidney function and increased numbers of bone marrow-derived endothelial cells | Mouse | [26] |
| miR-709, miR-217 and miR-696 | / | Plasma | Biomarker of AKI | Mouse | [27] |
| miR-10a | / | Plasma | Biomarker of AKI | Rat | [28] |
| miR-21 and miR-320 | / | Plasma | Biomarker of AKI | Rat | [30] |
| miR-200a | / | Plasma | Biomarker of AKI | Mouse | [31] |
| miR-210 | / | Plasma | Biomarker of AKI | Human | [32] |
| miR-210 | / | Urine | Biomarker of acute T cell-mediated rejection of renal allografts | Human | [33] |
AKI = acute kidney injury; EPC = endothelial progenitor cells; H2A.X = H2A histone family member X; MV = microvesicles
The majority of the human genome is transcribed into RNA transcripts with little or no protein coding potential. In fact, more than 98% are so called noncoding RNAs (ncRNAs), which are arbitrarily separated into long ncRNAs (lncRNAs, ≥200 nucleotides) and small ncRNAs (≤200 nucleotides) [7]. Information regarding the function of lncRNAs, as opposed to miRNAs, is largely lacking. However, it is now widely accepted that lncRNAs have major roles in epigenetic processes and cell function in the developmental phase, as well as during disease [34]. LncRNAs are novel regulatory RNA species, which may function as master regulators modifying the expression of mRNAs and miRNAs, and altering chromatin architecture. LncRNAs may either be nuclear-enriched or primarily found in the cytosol. Nuclear lncRNAs may associate with the chromatin architecture of genes on the same chromosome (in cis) or on another chromosome (in trans) [34]. Cytoplasmic lncRNAs may function as competing endogenous RNAs and thereby regulate the function of miRNAs [34]. A functional annotation has not been forwarded yet. This has led to the categorisation of lncRNAs on the basis of their location in the genome in relation to protein-coding genes. LncRNAs may therefore be viewed as: sense, antisense, bidirectional promoter-associated, intronic, intergenic and enhancer-associated [34]. In view of the lack of a functional annotation, Howard Chang and co-workers have proposed a mechanistic model regarding the role of lncRNAs [34, 35]. According to this suggested classification, lncRNAs are classified as: (1) signal lncRNAs, which are spatiotemporally transcribed in response to developmental cues, cellular context or diverse stimuli; (2) decoy lncRNAs, which impact on transcriptional regulation by titrating away transcription factors and other proteins from chromatin; (3) guide lncRNAs, which sequester ribonucleoprotein complexes and direct them to chromatin and other specific targets; and (4) scaffold lncRNAs, which interact with multiple partners to form a chromatin modifying complex (see fig. 2) [34, 35]. To date, the roles of a small number of lncRNAs have been characterised in more detail. The long intergenic RNA (lincRNA) Air has been shown to regulate imprinting, whereas X-chromosome inactivation has been shown to be influenced by Xist (X-inactive specific transcript) [34]. The lncRNA H19 has a tumour-suppressive effect and is imprinted with maternal expression [34]
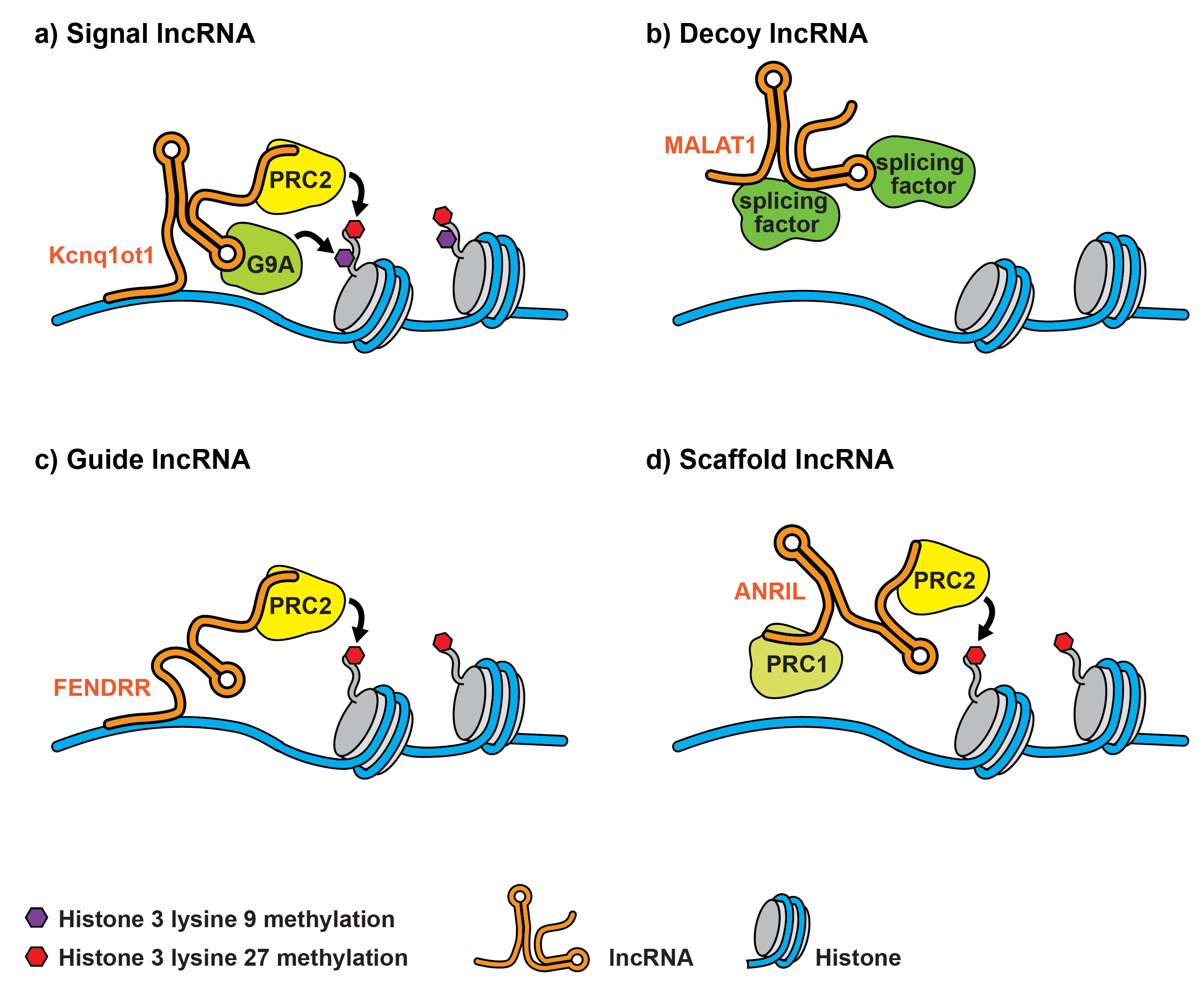
Figure 2 Mechanistic classification of lncRNAs as signal lncRNAs, which are spatiotemporally transcribed (a), decoy lncRNAs, which impact on transcriptional regulation by titrating away transcription factors (b), guide lncRNAs, which sequester ribonucleoprotein complexes and direct them to chromatin and other specific targets (c) and scaffold lncRNAs, which interact with multiple partners to form a chromatin modifying complex (d). Kcnq1ot1 = potassium voltage-gated channel subfamily Q member 1 antisense transcript 1; MALAT1 = metastasis-associated lung adenocarcinoma transcript 1; FENDRR = Fox F1 adjacent non-coding developmental regulatory RNA; PRC1 = polycomb repressive complex 1; PRC2 = polycomb repressive complex 2
Recently, the hypoxic and inflammatory dysregulation pattern of lncRNAs in cultured human proximal tubular epithelial cells was investigated [36]. The proximal tubular epithelial cells were exposed to hypoxia and treated with a cytokine cocktail implicated in clinical AKI (interleukin-6 [IL-6], tumour necrosis factor-α [TNF-α], and interferon-γ). A total number of 667 annotated lncRNAs were found, but only 14 different lncRNAs overlapped between treatment conditions. Hypoxia-sensitive lncRNAs were located in the vicinity of protein-coding genes, including those with roles in the Wnt/β-catenin signalling pathway, ATP metabolism and vitamin D receptor activation, as well as cellular differentiation. MIR210HG, linc-ATP13A4-8 and linc-KIAA1737-2 were amongst the most highly upregulated transcripts. These were detectable in human kidney tissue as well as microdissected proximal tubules from kidney transplant recipients. Addition of blood samples of sepsis patients to cultured cells induced expression of all investigated lncRNAs.
The lncRNA RANTES (regulated on activation, normal T-cell expressed and secreted) is induced in mice with I/R injury [37]. Kidney dysfunction, renal inflammation and tissue injury in RANTES knockout mice were shown to be ameliorated following renal I/R injury. Nuclear factor-κB (NF-κB) was transcriptionally activated RANTES. Moreover, a HIF-1α-regulated lncRNA, termed PRINS (psoriasis susceptibility-related RNA gene induced by stress), was proposed to interact with RANTES under hypoxic conditions. In a mouse model of unilateral ureteral obstruction, a large number of dysregulated lncRNAs was recently identified by means of RNA sequencing [38]. The antisense lncRNA Arid2-IR was found to be differentially regulated and associated with progressive renal inflammation in a unilateral ureteral obstruction model involving Smad3-knockout and wild-type littermate control animals [39]. Recently, our group analysed the contribution of the hypoxia-regulated lncRNA metastasis associated lung adenocarcinoma transcript 1 (Malat1) to ischaemic AKI [40]. Malat1 was highly induced in kidney biopsies and plasma of patients with AKI as well as in a mouse model of renal I/R injury. In addition, its expression increased in ex vivo sorted hypoxic endothelial and proximal tubular epithelial cells, as well as in cultured endothelial and proximal tubular epithelial cells. HIF-1α functioned as a transcriptional activator of Malat1. Malat1 silencing impaired the proliferation rate and reduced the number of ECs in the S-phase of the cell cycle. In vivo, Malat1 knockout and wild-type mice did not exhibit differences in the levels of renal injury, capillary rarefaction and fibrosis as well as survival rate and kidney function. RNA sequencing did not reveal differences in the expression of small RNAs and mRNAs between groups. LncRNA activated by transforming growth factor-β (TGF-β) (lncRNA-ATB) is highly upregulated in kidney biopsies of patients with acute renal allograft rejection [41].
Our group previously reported an initial study regarding the release and biomarker potential of lncRNAs in patients with AKI. Here, the intronic antisense lncRNA transcript predicting survival in AKI (TapsAKI, alternative nomenclature: MGAT3AS1) was shown to be massively increased in blood and kidney biopsies of patients with AKI [42]. TapsAKI proved to be an independent and powerful predictor of 28-day survival [42]. TapsAKI may be released from endothelial and tubular epithelial cells under stress conditions, since its expression level was increased by hypoxia or chemical anoxia (ATP depletion).
We subsequently investigated kidney biopsies and urine of kidney transplant patients with acute T cell-mediated renal allograft rejection. Three intergenic lncRNAs were identified: LNC-MYH13-3:1, RP11-395P13.3-001 and RP11-354P17.15-001 [43]. RP11-354P17.15-001 predicted acute rejection and loss of kidney function at 1 year after transplantation. Exposure of cultured tubular epithelial cells to the inflammatory cytokine IL-6, increased the levels of all lncRNAs, but only RP11-395P13.3-001 and RP11-354P17.15-001 expression increased in the cell culture supernatant, indicating that these lncRNAs might be secreted under inflammatory conditions.
LncRNAs with roles in rodent and human models of AKI are summarised in table 2.
Table 2 LncRNAs, their targets and pathophysiological effects in acute kidney injury.
| LncRNA | Target | Annotation | Pathophysiological effects | Organism | Reference |
|---|---|---|---|---|---|
| MIR210HG | Unknown | Intergenic | Induced by hypoxia and cytokine treatment in cultured proximal tubular epithelial cells | Human | [36] |
| linc-ATP13A4-8 | Unknown | Intergenic | Induced by hypoxia and cytokine treatment in cultured proximal tubular epithelial cells | Human | [36] |
| linc-KIAA1737-2 | Unknown | Intergenic | Induced by hypoxia and cytokine treatment in cultured proximal tubular epithelial cells | Human | [36] |
| PRINS | HIF-1α | Intronic | induced in hypoxia, regulated in RANTES knockout mice | Mouse | [37] |
| Arid2-IR | NF-κB, IL-1β | Intronic | Inflammation and fibrosis | Mouse | [39] |
| MALAT1 | Unknown | Intergenic | ischaemia/reperfusion-injury with lack of effect | Mouse | [40] |
| TapsAKI | Unknown | Antisense | Biomarker (plasma) in acute kidney injury | Human | [42] |
| RP11-354P17.15-001 | Unknown | Intergenic | Biomarker (urine) of acute T cell-mediated rejection of renal allografts | Human | [43] |
Arid2-IR = AT-rich interactive domain-containing protein 2 intronic region; HIF-1α = ; IL-1β = interleukin-1β; MALAT1 = metastasis associated lung adenocarcinoma transcript 1; NF-κB = nuclear factor-κB; PRINS = psoriasis susceptibility-related RNA gene induced by stress; RANTES = regulated on activation, normal T cell expressed and secreted; TapsAKI = transcript predicting survival in AKI
Circular RNAs (circRNAs) are suggested to be part of a so-called competing endogenous RNA class and may function as miRNA sponge transcripts. They are ubiquitously distributed and have diverse functions. CircRNAs are formed as single-stranded, circular molecules, in which the 3′ and 5′ ends are covalently linked [44–46]. The majority of circRNAs are generated by a process termed back-splicing. Here, a splice donor is joined with an upstream splice acceptor [45, 46]. They are ∼100 nucleotides in length and are highly present in the eukaryotic transcriptome and abundant in exosomes [47]. “Exonic” circRNAs (formed from exons) can be differentiated from “intronic” circRNAs, which contain a 2′-5′ carbon linkage at the branch point stemming from introns. It is currently believed that circRNAs are not translated into protein, since they are lacking a 5′ cap [48]. A characteristic feature of circRNAs is the “head-to-tail” splice junction, where exons are organised in reverse order compared with their chromosomal localisation. This is a consequence of the backsplicing reaction.
The RNA binding proteins Muscleblind [45], Quaking [49] or RNA binding motif protein 20 [50] may contribute to circRNA biogenesis. On the other hand, the RNA-editing enzyme adenosine deaminase RNA specific (ADAR1) blocks circRNAs biogenesis [51].
Platelets, neutrophils, B cells and haematopoietic stem cells may be important sources of circRNAs [52–54]. In addition, circRNAs may be actively secreted and detected in exosomes or small vesicles [55].
Recently, it was shown that circRNAs may function as remarkably stable biomarkers in human blood as a result of their resistance to exonucleases through circularisation [56]. Several studies have highlighted their biomarker potential in patients with atherosclerosis [57], disorders of the central nervous system [58], degenerative diseases [45], and cancers [55, 59].
The literature on the role of circRNAs in the kidney and especially AKI is scarce. An initial study showed that circRNAs may be important for renal development [60]. RNA sequencing confirmed that circRNAs are present and enriched in the kidney [61]. Several circRNAs have been shown to be altered in murine ischaemic AKI [62]. Our group has recently shown that the novel circRNA ciRS-126 predicts survival of critically ill patients with AKI [63]. We performed a global circRNA expression analysis using RNA isolated from blood of patients with AKI. This expression analysis yielded a number of dysregulated circRNAs. Circulating concentrations of three novel circRNAs, identified with array, were validated by means of quantitative polymerase chain-reaction tests (qPCR) in blood of patients with AKI. Circular RNA sponge of miR-126 (or ciRs-126) was the most altered compared with controls. CiRs-126 was shown to bioinformatically sponge miR-126-5p, which was found to be highly suppressed in AKI patients and hypoxic endothelial cells. Cox regression and Kaplan-Meier curve analysis revealed ciRs-126 to be an independent predictor of 28-day survival. Circulating concentrations of circRNAs, and more specifically ciRs-126, in patients with AKI may act as a predictor of mortality in this patient cohort.
Pharmacological treatment of patients with AKI is limited. A targeted and specific therapy in proximal tubular cells of the S3 segment or endothelial cells of the outer medulla, which are the primary target cells in ischaemic AKI, to halt or even reverse the progression of AKI development is largely lacking. Noncoding RNAs may be therapeutically used to ameliorate or retard disease progression, as recently shown by us regarding miR-21 silencing in murine diabetic nephropathy [64] and chronic allograft dysfunction following kidney transplantation [65], as well as miR-24 [66] inhibition regarding murine renal ischaemia reperfusion injury. MicroRNAs are highly conserved between different species and therefore hold huge potential as biomarkers and therapeutic targets in humans with kidney disease. LncRNAs are novel regulatory RNA species, which have been shown to critically impact on various cellular functions and may therefore be an ideal candidate for therapeutic interventions. In general, lncRNA conservation between species is low, thereby limiting their applicability for therapeutic intervention in humans following initial validation in animal studies. However, their functional and structural (secondary structure) homology might be much higher across species. Another obstacle is the fact that certain lncRNAs have several transcripts. Thus, the identification of specific mechanisms of action is not trivial.
The elucidation of the role of circRNAs is still in its infancy. It is currently unclear, how and to what extent they contribute to kidney disease. Future studies will aim to identify novel circRNAs with implications in kidney disease. Ultimately, modulation of noncoding RNAs may be a viable therapeutic option in the treatment of patients with kidney disease.
No financial support and no other potential conflict of interest relevant to this article was reported.
1 Barrantes F , Tian J , Vazquez R , Amoateng-Adjepong Y , Manthous CA . Acute kidney injury criteria predict outcomes of critically ill patients. Crit Care Med. 2008;36(5):1397–403. doi:.https://doi.org/10.1097/CCM.0b013e318168fbe0
2 Mehta RL , Kellum JA , Shah SV , Molitoris BA , Ronco C , Warnock DG , et al.; Acute Kidney Injury Network. Acute Kidney Injury Network: report of an initiative to improve outcomes in acute kidney injury. Crit Care. 2007;11(2):R31. doi:.https://doi.org/10.1186/cc5713
3 Uchino S , Kellum JA , Bellomo R , Doig GS , Morimatsu H , Morgera S , et al., Beginning and Ending Supportive Therapy for the Kidney (BEST Kidney) Investigators. Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA. 2005;294(7):813–8. doi:.https://doi.org/10.1001/jama.294.7.813
4 Kelly KJ . Acute renal failure: much more than a kidney disease. Semin Nephrol. 2006;26(2):105–13. doi:.https://doi.org/10.1016/j.semnephrol.2005.09.003
5 Bon D , Chatauret N , Giraud S , Thuillier R , Favreau F , Hauet T . New strategies to optimize kidney recovery and preservation in transplantation. Nat Rev Nephrol. 2012;8(6):339–47. doi:.https://doi.org/10.1038/nrneph.2012.83
6 Weight SC , Bell PR , Nicholson ML . Renal ischaemia--reperfusion injury. Br J Surg. 1996;83(2):162–70. doi:.https://doi.org/10.1002/bjs.1800830206
7 ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi:. https://doi.org/10.1038/nature11247
8 Djebali S , Davis CA , Merkel A , Dobin A , Lassmann T , Mortazavi A , et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi:.https://doi.org/10.1038/nature11233
9 Bonventre JV , Yang L . Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest. 2011;121(11):4210–21. doi:.https://doi.org/10.1172/JCI45161
10 Lorenzen JM , Batkai S , Thum T . Regulation of cardiac and renal ischemia-reperfusion injury by microRNAs. Free Radic Biol Med. 2013;64:78–84. doi:.https://doi.org/10.1016/j.freeradbiomed.2013.06.044
11 Lorenzen JM , Haller H , Thum T . MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 2011;7(5):286–94. doi:.https://doi.org/10.1038/nrneph.2011.26
12 Ambros V . A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57(1):49–57. doi:.https://doi.org/10.1016/0092-8674(89)90171-2
13 Chalfie M , Horvitz HR , Sulston JE . Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24(1):59–69. doi:.https://doi.org/10.1016/0092-8674(81)90501-8
14 Lim LP , Glasner ME , Yekta S , Burge CB , Bartel DP . Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi:.https://doi.org/10.1126/science.1080372
15 Lorenzen JM , Thum T . Circulating and urinary microRNAs in kidney disease. Clin J Am Soc Nephrol. 2012;7(9):1528–33. doi:.https://doi.org/10.2215/CJN.01170212
16 Wei Q , Bhatt K , He HZ , Mi QS , Haase VH , Dong Z . Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21(5):756–61. doi:.https://doi.org/10.1681/ASN.2009070718
17 Zhang L , Xu Y , Xue S , Wang X , Dai H , Qian J , et al. Implications of dynamic changes in miR-192 expression in ischemic acute kidney injury. Int Urol Nephrol. 2017;49(3):541–50. doi:.https://doi.org/10.1007/s11255-016-1485-7
18 Lorenzen JM , Kaucsar T , Schauerte C , Schmitt R , Rong S , Hübner A , et al. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J Am Soc Nephrol. 2014;25(12):2717–29. doi:.https://doi.org/10.1681/ASN.2013121329
19 Godwin JG , Ge X , Stephan K , Jurisch A , Tullius SG , Iacomini J . Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci USA. 2010;107(32):14339–44. doi:.https://doi.org/10.1073/pnas.0912701107
20 Shapiro MD , Bagley J , Latz J , Godwin JG , Ge X , Tullius SG , et al. MicroRNA expression data reveals a signature of kidney damage following ischemia reperfusion injury. PLoS One. 2011;6(8):e23011. doi:.https://doi.org/10.1371/journal.pone.0023011
21 Aguado-Fraile E , Ramos E , Sáenz-Morales D , Conde E , Blanco-Sánchez I , Stamatakis K , et al. miR-127 protects proximal tubule cells against ischemia/reperfusion: identification of kinesin family member 3B as miR-127 target. PLoS One. 2012;7(9):e44305. doi:.https://doi.org/10.1371/journal.pone.0044305
22 Cantaluppi V , Gatti S , Medica D , Figliolini F , Bruno S , Deregibus MC , et al. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 2012;82(4):412–27. doi:.https://doi.org/10.1038/ki.2012.105
23 Xu X , Kriegel AJ , Liu Y , Usa K , Mladinov D , Liu H , et al. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012;82(11):1167–75. doi:.https://doi.org/10.1038/ki.2012.241
24 Wilflingseder J , Sunzenauer J , Toronyi E , Heinzel A , Kainz A , Mayer B , et al. Molecular pathogenesis of post-transplant acute kidney injury: assessment of whole-genome mRNA and miRNA profiles. PLoS One. 2014;9(8):e104164. doi:.https://doi.org/10.1371/journal.pone.0104164
25 Wilflingseder J , Jelencsics K , Bergmeister H , Sunzenauer J , Regele H , Eskandary F , et al. miR-182-5p Inhibition Ameliorates Ischemic Acute Kidney Injury. Am J Pathol. 2017;187(1):70–9. doi:.https://doi.org/10.1016/j.ajpath.2016.09.011
26 Bijkerk R , van Solingen C , de Boer HC , van der Pol P , Khairoun M , de Bruin RG , et al. Hematopoietic microRNA-126 protects against renal ischemia/reperfusion injury by promoting vascular integrity. J Am Soc Nephrol. 2014;25(8):1710–22. doi:.https://doi.org/10.1681/ASN.2013060640
27 Bellinger MA , Bean JS , Rader MA , Heinz-Taheny KM , Nunes JS , Haas JV , et al. Concordant changes of plasma and kidney microRNA in the early stages of acute kidney injury: time course in a mouse model of bilateral renal ischemia-reperfusion. PLoS One. 2014;9(4):e93297. doi:.https://doi.org/10.1371/journal.pone.0093297
28 Wang JF , Zha YF , Li HW , Wang F , Bian Q , Lai XL , et al. Screening plasma miRNAs as biomarkers for renal ischemia-reperfusion injury in rats. Med Sci Monit. 2014;20:283–9. doi:.https://doi.org/10.12659/MSM.889937
29 Yu L , Gu T , Shi E , Wang Y , Fang Q , Wang C . Dysregulation of renal microRNA expression after deep hypothermic circulatory arrest in rats. Eur J Cardiothorac Surg. 2016;49(6):1725–31. doi:.https://doi.org/10.1093/ejcts/ezv460
30 Saikumar J , Hoffmann D , Kim TM , Gonzalez VR , Zhang Q , Goering PL , et al. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol Sci. 2012;129(2):256–67. doi:.https://doi.org/10.1093/toxsci/kfs210
31 Kito N , Endo K , Ikesue M , Weng H , Iwai N . miRNA Profiles of Tubular Cells: Diagnosis of Kidney Injury. BioMed Res Int. 2015;2015:465479. doi:.https://doi.org/10.1155/2015/465479
32 Lorenzen JM , Kielstein JT , Hafer C , Gupta SK , Kümpers P , Faulhaber-Walter R , et al. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol. 2011;6(7):1540–6. doi:.https://doi.org/10.2215/CJN.00430111
33 Lorenzen JM , Volkmann I , Fiedler J , Schmidt M , Scheffner I , Haller H , et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011;11(10):2221–7. doi:.https://doi.org/10.1111/j.1600-6143.2011.03679.x
34 Lorenzen JM , Thum T . Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12(6):360–73. doi:.https://doi.org/10.1038/nrneph.2016.51
35 Wang KC , Chang HY . Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14. doi:.https://doi.org/10.1016/j.molcel.2011.08.018
36 Lin J , Zhang X , Xue C , Zhang H , Shashaty MG , Gosai SJ , et al. The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. Am J Physiol Renal Physiol. 2015;309(11):F901–13. doi:.https://doi.org/10.1152/ajprenal.00290.2015
37 Yu TM , Palanisamy K , Sun KT , Day YJ , Shu KH , Wang IK , et al. RANTES mediates kidney ischemia reperfusion injury through a possible role of HIF-1α and LncRNA PRINS. Sci Rep. 2016;6(1):18424. doi:.https://doi.org/10.1038/srep18424
38 Arvaniti E , Moulos P , Vakrakou A , Chatziantoniou C , Chadjichristos C , Kavvadas P , et al. Whole-transcriptome analysis of UUO mouse model of renal fibrosis reveals new molecular players in kidney diseases. Sci Rep. 2016;6(1):26235. doi:.https://doi.org/10.1038/srep26235
39 Zhou Q , Huang XR , Yu J , Yu X , Lan HY . Long Noncoding RNA Arid2-IR Is a Novel Therapeutic Target for Renal Inflammation. Mol Ther. 2015;23(6):1034–43. doi:.https://doi.org/10.1038/mt.2015.31
40 Kölling M , Genschel C , Kaucsar T , Hübner A , Rong S , Schmitt R , et al. Hypoxia-induced long non-coding RNA Malat1 is dispensable for renal ischemia/reperfusion-injury. Sci Rep. 2018;8(1):3438. doi:.https://doi.org/10.1038/s41598-018-21720-3
41 Qiu J , Chen Y , Huang G , Zhang Z , Chen L , Na N . Transforming growth factor-β activated long non-coding RNA ATB plays an important role in acute rejection of renal allografts and may impacts the postoperative pharmaceutical immunosuppression therapy. Nephrology (Carlton). 2017;22(10):796–803. doi:.https://doi.org/10.1111/nep.12851
42 Lorenzen JM , Schauerte C , Kielstein JT , Hübner A , Martino F , Fiedler J , et al. Circulating long noncoding RNATapSaki is a predictor of mortality in critically ill patients with acute kidney injury. Clin Chem. 2015;61(1):191–201. doi:.https://doi.org/10.1373/clinchem.2014.230359
43 Lorenzen JM , Schauerte C , Kölling M , Hübner A , Knapp M , Haller H , et al. Long Noncoding RNAs in Urine Are Detectable and May Enable Early Detection of Acute T Cell-Mediated Rejection of Renal Allografts. Clin Chem. 2015;61(12):1505–14. doi:.https://doi.org/10.1373/clinchem.2015.243600
44 Jeck WR , Sharpless NE . Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61. doi:.https://doi.org/10.1038/nbt.2890
45 Ashwal-Fluss R , Meyer M , Pamudurti NR , Ivanov A , Bartok O , Hanan M , et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:.https://doi.org/10.1016/j.molcel.2014.08.019
46 Starke S , Jost I , Rossbach O , Schneider T , Schreiner S , Hung LH , et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–11. doi:.https://doi.org/10.1016/j.celrep.2014.12.002
47 Memczak S , Jens M , Elefsinioti A , Torti F , Krueger J , Rybak A , et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. doi:.https://doi.org/10.1038/nature11928
48 Li XF , Lytton J . A circularized sodium-calcium exchanger exon 2 transcript. J Biol Chem. 1999;274(12):8153–60. doi:.https://doi.org/10.1074/jbc.274.12.8153
49 Conn SJ , Pillman KA , Toubia J , Conn VM , Salmanidis M , Phillips CA , et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–34. doi:.https://doi.org/10.1016/j.cell.2015.02.014
50 Khan MA , Reckman YJ , Aufiero S , van den Hoogenhof MM , van der Made I , Beqqali A , et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ Res. 2016;119(9):996–1003. doi:.https://doi.org/10.1161/CIRCRESAHA.116.309568
51 Ivanov A , Memczak S , Wyler E , Torti F , Porath HT , Orejuela MR , et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–7. doi:.https://doi.org/10.1016/j.celrep.2014.12.019
52 Salzman J , Gawad C , Wang PL , Lacayo N , Brown PO . Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi:.https://doi.org/10.1371/journal.pone.0030733
53 Alhasan AA , Izuogu OG , Al-Balool HH , Steyn JS , Evans A , Colzani M , et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016;127(9):e1–11. doi:.https://doi.org/10.1182/blood-2015-06-649434
54 Preußer C , Hung LH , Schneider T , Schreiner S , Hardt M , Moebus A , et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. 2018;7(1):1424473. doi:.https://doi.org/10.1080/20013078.2018.1424473
55 Li Y , Zheng Q , Bao C , Li S , Guo W , Zhao J , et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4. doi:.https://doi.org/10.1038/cr.2015.82
56 Memczak S , Papavasileiou P , Peters O , Rajewsky N . Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015;10(10):e0141214. doi:.https://doi.org/10.1371/journal.pone.0141214
57 Burd CE , Jeck WR , Liu Y , Sanoff HK , Wang Z , Sharpless NE . Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. doi:.https://doi.org/10.1371/journal.pgen.1001233
58 Lukiw WJ . Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. 2013;4:307. doi:.https://doi.org/10.3389/fgene.2013.00307
59 Li P , Chen S , Chen H , Mo X , Li T , Shao Y , et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–6. doi:.https://doi.org/10.1016/j.cca.2015.02.018
60 Chao CW , Chan DC , Kuo A , Leder P . The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4(9):614–28. doi:.https://doi.org/10.1007/BF03401761
61 Xu T , Wu J , Han P , Zhao Z , Song X . Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(S6, Suppl 6):680. doi:.https://doi.org/10.1186/s12864-017-4029-3
62 Zhou J , Chen H , Fan Y . Systematic analysis of the expression profile of non-coding RNAs involved in ischemia/reperfusion-induced acute kidney injury in mice using RNA sequencing. Oncotarget. 2017;8(59):100196–215. doi:.https://doi.org/10.18632/oncotarget.22130
63 Kölling M , Seeger H , Haddad G , Kistler A , Nowak A , Faulhaber-Walter R , et al. The circular RNA ciRs-126 predicts survival in critically ill patients with acute kidney injury. Kidney Int Rep. 2018;3(5):1144–52. doi:.https://doi.org/10.1016/j.ekir.2018.05.012
64 Kölling M , Kaucsar T , Schauerte C , Hübner A , Dettling A , Park JK , et al. Therapeutic miR-21 silencing ameliorates diabetic kidney disease in mice. Mol Ther. 2017;25(1):165–80. doi:.https://doi.org/10.1016/j.ymthe.2016.08.001
65 Schauerte C , Hübner A , Rong S , Wang S , Shushakova N , Mengel M , et al. Antagonism of profibrotic microRNA-21 improves outcome of murine chronic renal allograft dysfunction. Kidney Int. 2017;92(3):646–56. doi:.https://doi.org/10.1016/j.kint.2017.02.012
66 Lorenzen JM , Kaucsar T , Schauerte C , Schmitt R , Rong S , Hübner A , et al. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J Am Soc Nephrol. 2014;25(12):2717–29. doi:.https://doi.org/10.1681/ASN.2013121329
No financial support and no other potential conflict of interest relevant to this article was reported.