A community outbreak of Legionnaires’ disease in Geneva, Switzerland, June to September 2017
DOI: https://doi.org/10.4414/smw.2018.14687
Marie-Céline
Zanellaa, Sabine
Yerlya, Abdessalam
Cherkaouib, Gesuele
Renzib, Aline
Mamina, Laura Lourenço
Cordesc, Elisabeth
Delaported, Zofia
Baranczuk-Turskaef, Olivia
Keiserf, Jacques
Schrenzelb, Stephan
Harbarthg, Valeria
Gaiah, Laurent
Kaisera
aLaboratory of Virology, Division of Laboratory Medicine and Division of Infectious Diseases, University of Geneva Hospitals, Switzerland; University of Geneva Medical School, Switzerland
bLaboratory of Bacteriology, Division of Laboratory Medicine and Division of Infectious Diseases, Geneva University Hospitals, Switzerland; University of
Geneva Medical School, Switzerland
cUniversity of Geneva Medical School, Switzerland
dCantonal Health Service, General Directorate for Health, Geneva, Switzerland
eUniversity of Geneva, Institute of Global Health, Geneva, Switzerland
fInstitute of Mathematics, University of Zurich, Switzerland
gInfection Control Programme and WHO Collaborating Centre on Patient Safety, The University of Geneva Hospitals and Faculty of Medicine, Geneva, Switzerland
hSwiss National Reference Centre for Legionella
Summary
PURPOSE
Eight confirmed cases of Legionnaires’ disease were identified at the Geneva University Hospitals between 28 July 2017 and 02 August 2017, leading to a detailed outbreak investigation.
METHODS
Legionnaires’ disease cases were defined according to Swiss and European (ELDSNet) consensus guidelines. An outbreak investigation task force was put in place. Patients were interviewed, when feasible, with a standard questionnaire. A Legionella pneumophila urinary antigen test was performed in all cases. Lower respiratory tract (LRT) specimens were collected for culture, polymerase chain-reaction (PCR) assay, monoclonal antibody subtyping and sequenced-based typing (SBT). Multiple environmental samples were collected. Case geographical mapping was performed and local meteorological data were obtained.
RESULTS
Thirty-four confirmed cases of Legionnaires’ disease were identified between 20 June 2017 and 16 September 2017, including 28 patients living in the Canton of Geneva and 6 cases in neighbouring cantons and France. The case fatality rate was 8.8%. The urinary antigen test was positive in 32/34 (94.1%) cases. Among the 17/34 (50%) cases with available LRT specimens, 8 (47.1%) were culture/PCR positive, 5 (29.4%) were PCR positive only, and 4 (23.5%) were culture/PCR negative. Monoclonal antibody subtyping and SBT on 12 samples allowed subtype identification of 8 samples, with a predominance of L. pneumophila serogroup-1 subtype-France/Allentown ST23 among clinical isolates. A specific city area was identified as a possible outbreak epicentre in 25/34 (73.5%) cases, although molecular analysis of clinical and environmental specimens revealed heterogeneous subtypes of L. pneumophila.
CONCLUSIONS
In this largest documented outbreak of Legionnaires’ disease in Switzerland, we report prompt outbreak identification, leading to timely initiation of a detailed, well-orchestrated clinical and epidemiological investigation.
Introduction
Legionella species are rod-shaped gram-negative bacteria ubiquitously found in freshwater environments and man-made environments such as hot water systems and cooling towers and are the causative agents of Legionnaires’ disease, a disease named after the first documented outbreak, during an American Legion Convention in 1976 [1, 2]. Legionnaires’ disease results mainly from the inhalation of aerosols containing the bacterium Legionella, and less commonly from the aspiration of drinking water [3]. Legionella is not usually transmitted from person to person and only one probable person-to-person transmission has been reported [4]. Legionnaires’ disease represents 2 to 20% of community-acquired pneumonia cases [5]. The majority of cases are reported as isolated and sporadic cases, and clusters (cases associated in space and time) and outbreaks (clusters with a suspected common source) can also occur. Among the 60 species and more than 70 serogroups of the genus Legionella, the majority of cases of Legionnaires’ disease in Europe and the United States are caused by L. pneumophila, and especially the L. pneumophila serogroup 1 [6]. Beyond the screening and diagnosis of Legionnaires’ disease cases that can be performed with urinary antigen tests, the identification of Legionella species, serogroups and subtypes in clinical and environmental samples should be performed with monoclonal antibody subtyping (mAbs) and sequence-based typing (SBT), which are of particular importance for epidemiological purposes.
We describe here the largest documented outbreak of Legionnaires’ disease in Switzerland, which occurred in the Canton of Geneva between 20 June and 12 September 12 2017. The outbreak of pneumonia was detected on 2 August 2 2017 when the Geneva University Hospitals (HUG) notified the Public Health Service of Geneva of eight positive Legionella urinary antigen tests from patients with pneumonia, all of whom resided in the Canton of Geneva, over the period of 28 July to 2 August. In contrast, a mean of 19.4 cases per year (range 16–26) were recorded in Geneva between 2012 and 2016. The HUG and the Public Health Service convened an outbreak control team on 2 August to conduct investigations and control the outbreak of Legionnaires’ disease.
Materials and methods
An active surveillance system to identify cases of Legionnaires’ disease was implemented in the HUG and information letters were sent to all general practitioners and clinics in Geneva. The Public Health Service reviewed cases reported in Geneva since 1 May 2017 (in Switzerland, cases of Legionnaires’ disease are mandatorily notified) and searched for cases associated with travel to Geneva through the European Legionnaires’ Disease Surveillance Network (ELDSNet).
Case definitions
Legionnaires’ disease cases were defined according to the Swiss and European (ELDSNet) case definition [7, 8]. Probable cases were defined as individuals meeting clinical (pneumonia) and epidemiological (environmental exposure to the Canton of Geneva with a history of living, working or visiting Geneva from 1 May 2017) criteria, and at least one microbiological criterion for a probable case. Confirmed cases meet clinical, epidemiological and at least one microbiological criterion for a confirmed case. Cases were classified as travel-associated if the individual stayed in accommodation away from home 2 to 10 days prior to developing symptoms. Cases were classified as nosocomial if they stayed in a hospital or healthcare facility 2 to 10 days prior to developing symptoms.
Epidemiological investigations
All cases (and/or their relatives) were interviewed by three dedicated investigators with a standard questionnaire comprising details of residential and working addresses, occupation, transportation and places visited (duration and frequency of visits) in the two weeks preceding symptom onset. The geographical distribution of home addresses was determined using R (R-3.3.3, ggmap, ggplot2).
Environmental investigations
The Public Health Service and the Official Food and Veterinary Control Authority of Geneva conducted environmental investigations. Possible airborne sources (including declared cooling towers of wet cooling systems) in Geneva were mapped and inspected, and water samples were collected for microbiological analyses. Meteorological data in Geneva from 1 June to 31 August 2017, in particular temperature and humidity data, were obtained through the Federal Office of Meteorology and Climatology MeteoSwiss [9].
Laboratory investigations
During the active surveillance period, from 2–18 August, clinicians in the HUG were asked to perform, whenever possible, (on all suspected Legionnaires’ disease cases) an L. pneumophila urinary rapid antigen test (Sofia Quidel, San Diego, CA, USA) and additionally, to collect lower respiratory tract specimens for systematic culture on standard and selective media (BCYE and BMPA, Oxoid), and for Legionella polymerase chain-reaction (PCR) assays (FTD Atypical CAP, Fast-Track Diagnostics, Luxembourg).
To compare the performance of the L. pneumophila urinary antigen rapid tests, all available -20°C stored urine samples collected during the outbreak were analysed with a BinaxNow assay (Alere). The tests were performed according to the manufacturers’ instructions.
Clinical and environmental isolates were then sent to the Reference Centre for Legionella in Bellinzona for monoclonal antibody subtyping (mAbs) and sequenced-based typing (SBT). For culture-negative but PCR-positive clinical samples, extracted DNA samples were also sent to the Reference Centre for further analysis. The Dresden Panel [10] was used for mAbs; SBT [11, 12] and nested-SBT [13, 14] were performed as previously described.
Results
Descriptive epidemiology
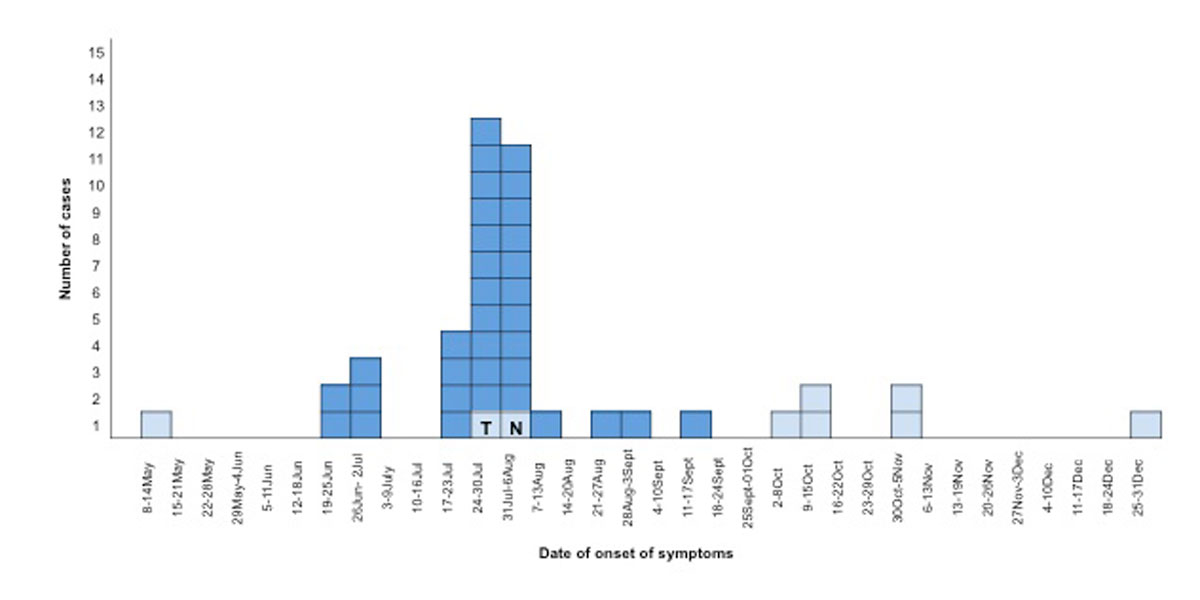
The review of previous notifications starting on 1 May 2017, as well as case tracing in the areas neighbouring Geneva, allowed the identification of 34 cases overall. Based on the epidemic curve (fig. 1), the outbreak started on the third week of June 2017 (the first patient presented with onset of symptoms on 20 June 20) and reached a peak between 25 July and 6 August. The outbreak ended in the third week of September (the last case presented with onset of symptoms on 12 September and diagnosis was confirmed on 16 September) (fig. 1). The epidemic curve (fig. 1) demonstrates that in the last week of September, the incidence returned to baseline (considering a mean of 1.62 cases, range: 1.33–2.16 cases, notified per month between 2012 and 2016 in Geneva). We thus consider the end of that month as the point at which the outbreak ended.
The Geneva community outbreak resulted in 34 confirmed Legionnaires’ disease cases, including 28 patients living in the Canton of Geneva and 6 cases in neighbouring France or other cantons, but all with an epidemiological exposure to Geneva. The median age of patients was 62 years (range 18–97), 76.5% (n = 26) were males, and the overall case-fatality rate was 8.8% (3/34). Eighteen patients (52.9%) were smokers or ex-smokers and nine (26.5%) had no co-morbidity. All confirmed cases received macrolide- or quinolone-based therapy. For the 26/34 (76.5%) patients with completely available medical information, characteristics are summarised in table 1.
Table 1 Clinical characteristics of confirmed cases of Legionnaires’ disease with completely available medical information (n = 26).
|
Characteristics
|
Number of patients (%)
|
| University Hospitals of Geneva admission |
24 (92.3%) |
| Intensive care unit admission |
5 (19.2%) |
| Case fatality rate |
3 (11.5%) |
|
General characteristics
|
|
| Chronic lung disease (asthma, COPD, OSAS, restrictive syndrome, lung cancer) |
9 (34.6%) |
| Immunosuppressive therapy |
3 (11.5%) |
|
Clinical manifestations
|
|
|
Respiratory symptoms
|
26 (100%) |
| Gastro-intestinal symptoms |
12 (46.1%) |
| Neurological symptoms |
9 (34.6%) |
| Pneumonia |
26 (100%) |
|
Laboratory
|
|
| CRP, median (range) |
298 mg/l (14–531 mg/l) |
| White blood cell count, median (range) |
11.6 G/l (3.4–32.3 G/l) |
| Natraemia, median (range) |
133 mmol/l (115–146 mmol/l) |
During the outbreak period, we also identified one case as travel-associated to Spain (deceased patient) and one as a possible healthcare-acquired case (fig. 1). No cases of Legionnaires’ disease associated with travel to Geneva was reported by ELDSNet.
The evaluation of the meteorological conditions from Geneva showed that the mean temperatures were on average 3.2, 1.2 and 1.7°C above the climatological mean (1981–2010) during June, July and August, respectively, in association with stormy conditions in late July and early August. A heat wave was recorded in the Geneva area from 19–24 June, with the third highest mean temperature recorded since 1864.
Laboratory investigations
Thirty-two of the 34 (94.1%) cases were urinary antigen test positive and two cases were antigen negative but culture and PCR positive. Culture and PCR assays were performed on respiratory tract specimens of 126 and 84 HUG suspected cases respectively as part of the large systematic screening implemented. Among the 34 cases, 17 did not have an available respiratory specimen. Among the 17 cases with available respiratory specimens, 8 (47.1%) were culture and PCR positive and 5 (29.4%) were PCR positive only. PCR and culture were negative for 4 cases (23.5%).
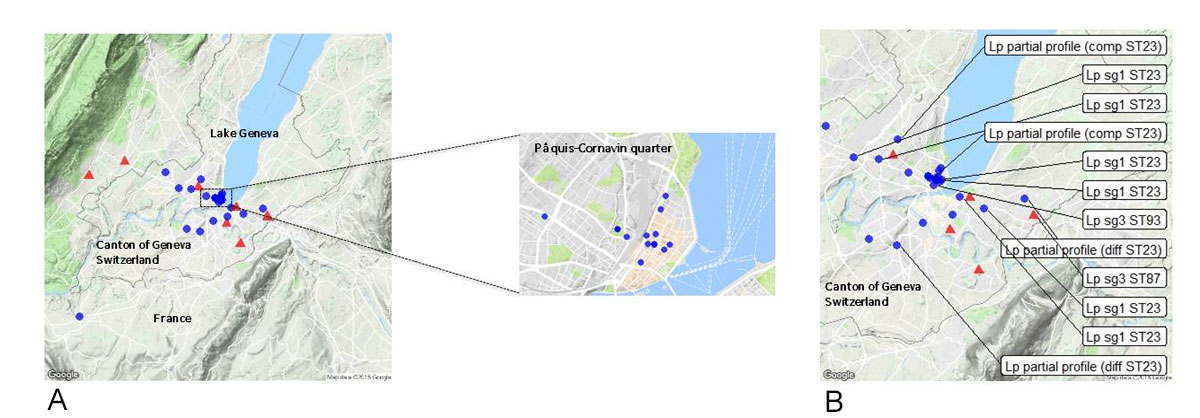
mAbs and SBT analyses were performed on 12 samples (7 culture positive, and 5 culture negative but PCR positive). These analyses allowed the complete subtype identification for 8 samples: L. pneumophila serogroup 1 subtype France/Allentown ST23 (6 samples) and L. pneumophila serogroup 3 subtype ST87 (one case, negative urinary antigen test) and ST93 (one case, positive urinary antigen test). SBT analysis resulted in partial identification for four samples: two partial profiles were compatible with ST23 alleles (flaA 2 and neuA 6) and two partial profiles were different from ST23. All patients infected with L. pneumophila subgroup 1 subtype France/Allentown ST23, as well as the one patient with serogroup 3 ST93 and the two patients with partly identified subtypes compatible with ST23, had an exposure to the Pâquis-Cornavin area in Geneva (fig. 2).
Comparison of two urinary antigen rapid tests
Of the 38 available urine samples which were positive with the Sofia assay, 29 (76%) were positive with the BinaxNow assay, 2 samples were invalid (control line absent) and 7 samples were negative (table 2). All negative samples with the Sofia assay were negative with the BinaxNow. Repetition of the discordant results yielded similar results for the Sofia assay, whereas three samples were found positive with the BinaxNow assay (two invalid and one negative results) (table 2).
Table 2 Comparison of two L. pneumophila urinary antigen rapid tests, Sofia and BinaxNow, on 38 available urine samples.
|
Sofia
|
| Positive |
Negative |
|
BinaxNow
|
Positive |
29 |
0 |
| Undetermined |
2 |
0 |
| Negative |
7 |
32 |
Outbreak management
Case interviews identified overlapping locations and local travel routes pointing to the Pâquis-Cornavin city area for 25/34 (73.5%) cases, comprising 13 residents, 2 workers and 10 visitors (people attending cafés, restaurants, shops or having regular activities in the area for more than two hours per day) (fig. 2). Measures were thus first taken in this area from 9–11 August, with sampling in 12 cooling towers at 7 sites, followed by disinfection procedures on 11 August. The sampled cultures of two cooling towers revealed concentrations of L. pneumophila (1.5×106 CFU/l and 3×103 CFU/l in the samples of cooling towers numbers 1 and 2, respectively) above the threshold at which resampling, maintenance and disinfection procedures are recommended by Swiss regulations [8]. mAbs and SBT of several colonies isolated from the two environmental samples revealed the presence of L. pneumophila serogroup 1 subgroup OLDA ST1, discordant from the clinical subtypes (12 morphologically different colonies from samples of cooling tower number 1 and the only three colonies from samples of cooling tower number 2 were analysed after isolation on GVPC (Biomerieux) specific culture media and thermic treatment). Water sampling of Lake Geneva and the drinking water network did not reveal concentrations of Legionella spp. above recommended limits and thus no specific measures were implemented for the use of these water sources.
Discussion
According to the Swiss Federal Office of Public Health, this was the largest documented community-associated outbreak of Legionnaires’ disease in Switzerland to date, with a total of 34 confirmed cases attributed to the outbreak.
Case interviews provided information for the rapid identification of a possible source area at Pâquis-Cornavin, a highly frequented area in which patients were possibly exposed to an environmental source, and in which investigations and control measures have been rapidly performed. All patients with L. pneumophila serogroup 1 ST23 had an exposure to the Pâquis-Cornavin area. Timely collection of lower respiratory tract specimens and environmental samplings did not allow the identification of a common source (e.g. cooling tower). The absence of an exhaustive register of at-risk water systems in Geneva could have contributed to these inconclusive results. The incidence of Legionnaires’ disease has been previously associated with specific meteorological conditions of high temperature and humidity [15–17]. Such weather conditions, reported in Geneva before and during this outbreak, could have contributed to the occurrence of Legionnaires’ disease cases. Outbreak control was achieved possibly by both the specific control measures implemented (enhanced disinfection of cooling towers in the Pâquis-Cornavin area) and changes in meteorological conditions (lower temperatures), or other unidentified reasons.
The use of PCR assays allowed identification of 2/2 (100%) of urinary antigen negative Legionnaires’ disease cases. Furthermore, partial and complete subtype profiles could be identified in 4/5 and 1/5 cases with culture negative samples respectively. Subtyping and SBT analysis of 12 clinical strains revealed at least 5 different subtypes: these results could reflect the diversity of Legionella subtypes among patients and in environmental sources, particularly after disinfection measures [1, 2]. The L. pneumophila serogroup 1 ST23 sequence type, which has been previously described in other outbreaks and sporadic community cases in Europe [18, 19], was the predominant subtype identified in clinical samples. L. pneumophila serogroup 3, a serogroup notably described in nosocomial cases and immunocompromised patients [20, 21], was identified in two clinical samples: one patient with idiopathic pulmonary fibrosis and another with lung cancer.
Whole genome sequencing (WGS) has recently been used in several studies with single nucleotide polymorphism (SNP) based, core-genome based and pan-genome based analysis and could provide better identification of clinical isolates of an outbreak and environmental isolates when compared to the gold standard SBT method [22–25]. The use of WGS on the clinical isolates of this outbreak could be of particular interest to obtain further clarification into the relatedness of cases.
Finally, our results confirmed that the Sofia test exhibits higher sensitivity than the BinaxNow, as reported in recent studies [26, 27].
A limitation of this study is the possible underestimation of cases, particularly among outpatients who possibly received adequate empiric treatment without a diagnostic test.
Conclusion
We report the largest community outbreak of Legionnaires’ disease in Switzerland with 34 confirmed cases, 24 hospitalised patients at HUG including 5 intensive care unit admissions and an overall mortality of 8.8%. Meteorological conditions may have contributed to the development and resolution of the outbreak. The collaborative work involving the HUG, Public Health Service and the Reference Centre for Legionella allowed for rapid investigations and the implementation of outbreak management and control measures. Interviews of cases allowed identification of a potential area of Geneva where infections occurred, but microbiological investigations could not identify a common source. Analyses of clinical samples revealed heterogeneous L. pneumophila subtypes with a predominance of serogroup 1 subtype France/Allentown ST23 among clinical samples. The urinary antigen test is an appropriate screening test, allowing identification of 94.1% of cases. This study highlights the importance of lower respiratory sample collection and the use of multiple diagnostic tools. In particular, the use of PCR with nested-SBT analysis to complement culture should be considered in future outbreaks of Legionnaires’ disease.
Acknowledgments
The authors would like to acknowledge the contribution of all members of the outbreak team, especially all members of the Laboratory of Virology and Bacteriology of Geneva University Hospitals, including Isabelle Arm-Vernez, Dr Pascal Cherpillod and Dr Samuel Cordey. We thank the staff of the Geneva University Hospitals and all members of the Division of Infectious Diseases and Infections Control Program of Geneva University Hospitals, including Dr Dionysios Neofytos for constructive comments and Elhadj Bah. We acknowledge Dr Barbara Bertisch and Professor Antoine Flahault from the Institute for Global Health for expert advice. We also thank Professor Jacques-André Romand for the coordination of the public health measures. Finally, we acknowledge Dr Patrick Edder and Nathalie Maury from the Official Food and Veterinary Control Authority of Geneva.
References
1
Fraser
DW
,
Tsai
TR
,
Orenstein
W
,
Parkin
WE
,
Beecham
HJ
,
Sharrar
RG
, et al.
Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297(22):1189–97. doi:.https://doi.org/10.1056/NEJM197712012972201
2
McDade
JE
,
Shepard
CC
,
Fraser
DW
,
Tsai
TR
,
Redus
MA
,
Dowdle
WR
. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297(22):1197–203. doi:.https://doi.org/10.1056/NEJM197712012972202
3
Fields
BS
,
Benson
RF
,
Besser
RE
. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15(3):506–26. doi:.https://doi.org/10.1128/CMR.15.3.506-526.2002
4
Correia
AM
,
Ferreira
JS
,
Borges
V
,
Nunes
A
,
Gomes
B
,
Capucho
R
, et al.
Probable Person-to-Person Transmission of Legionnaires’ Disease. N Engl J Med. 2016;374(5):497–8. doi:.https://doi.org/10.1056/NEJMc1505356
5
Torres
A
,
Blasi
F
,
Peetermans
WE
,
Viegi
G
,
Welte
T
. The aetiology and antibiotic management of community-acquired pneumonia in adults in Europe: a literature review. Eur J Clin Microbiol Infect Dis. 2014;33(7):1065–79. doi:.https://doi.org/10.1007/s10096-014-2067-1
6Bartram J, Chartier Y, Lee JV, Pond K, Surman-Lee S, eds. Legionella and the Prevention of Legionellosis. World Health Organization; 2007. Available at.
72008/426/EC: Commission Decision of 28 April 2008 amending Decision 2002/253/EC laying down case definitions for reporting communicable diseases to the Community network under Decision No 2119/98/EC of the European Parliament and of the Council. OJ L 159, 18.6.2008, p. 46–90.
8Federal Office of Public Health. Legionnella et légionnellose. 2009. https://www.bag.admin.ch/bag/fr/home/themen/mensch-gesundheit/uebertragbare-krankheiten/infektionskrankheiten-a-z/legionellose.html.
9
Federal Office of Meteorology and Climatology MeteoSwiss. Available from: http://www.meteoswiss.admin.ch.
10
Helbig
JH
,
Lück
PC
,
Knirel
YA
,
Witzleb
W
,
Zähringer
U
. Molecular characterization of a virulence-associated epitope on the lipopolysaccharide of Legionella pneumophila serogroup 1. Epidemiol Infect. 1995;115(1):71–8. doi:.https://doi.org/10.1017/S0950268800058131
11
Gaia
V
,
Fry
NK
,
Afshar
B
,
Lück
PC
,
Meugnier
H
,
Etienne
J
, et al.
Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol. 2005;43(5):2047–52. doi:.https://doi.org/10.1128/JCM.43.5.2047-2052.2005
12
Ratzow
S
,
Gaia
V
,
Helbig
JH
,
Fry
NK
,
Lück
PC
. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J Clin Microbiol. 2007;45(6):1965–8. doi:.https://doi.org/10.1128/JCM.00261-07
13
Ginevra
C
,
Lopez
M
,
Forey
F
,
Reyrolle
M
,
Meugnier
H
,
Vandenesch
F
, et al.
Evaluation of a nested-PCR-derived sequence-based typing method applied directly to respiratory samples from patients with Legionnaires’ disease. J Clin Microbiol. 2009;47(4):981–7. doi:.https://doi.org/10.1128/JCM.02071-08
14
Mentasti
M
,
Fry
NK
,
Afshar
B
,
Palepou-Foxley
C
,
Naik
FC
,
Harrison
TG
. Application of Legionella pneumophila-specific quantitative real-time PCR combined with direct amplification and sequence-based typing in the diagnosis and epidemiological investigation of Legionnaires’ disease. Eur J Clin Microbiol Infect Dis. 2012;31(8):2017–28. doi:.https://doi.org/10.1007/s10096-011-1535-0
15
Conza
L
,
Casati
S
,
Limoni
C
,
Gaia
V
. Meteorological factors and risk of community-acquired Legionnaires’ disease in Switzerland: an epidemiological study. BMJ Open. 2013;3(3):e002428. doi:.https://doi.org/10.1136/bmjopen-2012-002428
16
Fisman
DN
,
Lim
S
,
Wellenius
GA
,
Johnson
C
,
Britz
P
,
Gaskins
M
, et al.
It’s not the heat, it’s the humidity: wet weather increases legionellosis risk in the greater Philadelphia metropolitan area. J Infect Dis. 2005;192(12):2066–73. doi:.https://doi.org/10.1086/498248
17
Simmering
JE
,
Polgreen
LA
,
Hornick
DB
,
Sewell
DK
,
Polgreen
PM
. Weather-Dependent Risk for Legionnaires’ Disease, United States. Emerg Infect Dis. 2017;23(11):1843–51. doi:.https://doi.org/10.3201/eid2311.170137
18
Sánchez-Busó
L
,
Coscollà
M
,
Palero
F
,
Camaró
ML
,
Gimeno
A
,
Moreno
P
, et al.
Geographical and Temporal Structures of Legionella pneumophila Sequence Types in Comunitat Valenciana (Spain), 1998 to 2013. Appl Environ Microbiol. 2015;81(20):7106–13. doi:.https://doi.org/10.1128/AEM.02196-15
19
Fontana
S
,
Scaturro
M
,
Rota
MC
,
Caporali
MG
,
Ricci
ML
. Molecular typing of Legionella pneumophila serogroup 1 clinical strains isolated in Italy. Int J Med Microbiol. 2014;304(5-6):597–602. doi:.https://doi.org/10.1016/j.ijmm.2014.04.004
20
Oren
I
,
Zuckerman
T
,
Avivi
I
,
Finkelstein
R
,
Yigla
M
,
Rowe
JM
. Nosocomial outbreak of Legionella pneumophila serogroup 3 pneumonia in a new bone marrow transplant unit: evaluation, treatment and control. Bone Marrow Transplant. 2002;30(3):175–9. doi:.https://doi.org/10.1038/sj.bmt.1703628
21
Chien
ST
,
Fong
C-M
,
Hsueh
JC
,
Lee
T-M
,
Ben
R-J
,
Chou
S-T
, et al.
Epidemiological investigation of a case of nosocomial Legionnaires’ disease in Taiwan: implications for routine environmental surveillance. Clin Microbiol Infect. 2010;16(6):761–3. doi:.https://doi.org/10.1111/j.1469-0691.2009.02890.x
22
Timms
VJ
,
Rockett
R
,
Bachmann
NL
,
Martinez
E
,
Wang
Q
,
Chen
SC
, et al.
Genome Sequencing Links Persistent Outbreak of Legionellosis in Sydney (New South Wales, Australia) to an Emerging Clone of Legionella pneumophila Sequence Type 211. Appl Environ Microbiol. 2018;84(5):e02020-17. doi:.https://doi.org/10.1128/AEM.02020-17
23
Petzold
M
,
Prior
K
,
Moran-Gilad
J
,
Harmsen
D
,
Lück
C
. Epidemiological information is key when interpreting whole genome sequence data - lessons learned from a large Legionella pneumophila outbreak in Warstein, Germany, 2013. Euro Surveill. 2017;22(45). doi:.https://doi.org/10.2807/1560-7917.ES.2017.22.45.17-00137
24
David
S
,
Mentasti
M
,
Tewolde
R
,
Aslett
M
,
Harris
SR
,
Afshar
B
, et al.
Evaluation of an Optimal Epidemiological Typing Scheme for Legionella pneumophila with Whole-Genome Sequence Data Using Validation Guidelines. J Clin Microbiol. 2016;54(8):2135–48. doi:.https://doi.org/10.1128/JCM.00432-16
25
Schjørring
S
,
Stegger
M
,
Kjelsø
C
,
Lilje
B
,
Bangsborg
JM
,
Petersen
RF
, et al.; ESCMID Study Group for Legionella Infections (ESGLI). Genomic investigation of a suspected outbreak of Legionella pneumophila ST82 reveals undetected heterogeneity by the present gold-standard methods, Denmark, July to November 2014. Euro Surveill. 2017;22(25):30558. doi:.https://doi.org/10.2807/1560-7917.ES.2017.22.25.30558
26
Beraud
L
,
Gervasoni
K
,
Freydiere
AM
,
Descours
G
,
Ranc
AG
,
Vandenesch
F
, et al.
Comparison of Sofia Legionella FIA and BinaxNOW® Legionella urinary antigen card in two national reference centers. Eur J Clin Microbiol Infect Dis. 2015;34(9):1803–7. doi:.https://doi.org/10.1007/s10096-015-2415-9
27
Ranc
AG
,
Carpentier
M
,
Beraud
L
,
Descours
G
,
Ginevra
C
,
Maisonneuve
E
, et al.
Legionella pneumophila LPS to evaluate urinary antigen tests. Diagn Microbiol Infect Dis. 2017;89(2):89–91. doi:.https://doi.org/10.1016/j.diagmicrobio.2017.06.013