RNA metabolism in Staphylococcus aureus virulence
DOI: https://doi.org/10.4414/smw.2017.14527
Joshua
Armitano, Patrick
Linder
Dept. of Microbiology and Molecular Medicine, Faculty of Medicine, University of Geneva, Switzerland
Summary
The opportunistic pathogen Staphylococcus aureus encounters a variety of host defence systems depending on its localisation during colonisation in the nares, systemic infections within the body, or persistent infections within cells or embedded in biofilms. To respond rapidly to these different environments, this bacterium has evolved, in its longstanding interaction with animal and human hosts, a variety of mechanisms to fine-tune its gene expression. RNA metabolism, including transcription, processing, translation into proteins and RNA decay, is a central player in this response and might in the future be used to treat this feared pathogen.
Introduction
Staphylococcus aureus is an opportunistic pathogen that colonises the nares of 20 to 30% persons [1]. In most cases this harmless colonisation remains without consequences. But this commensal can also cause skin infections such as boils or carbuncles. More importantly, it can also cause severe or life-threatening infections such as bacteraemia, endocarditis, or osteomyelitis. Finally, several strains of S. aureus produce toxins that may cause food poisoning or lead to the feared toxic-shock syndrome caused by superantigens. S. aureus is notoriously known for healthcare-associated infections by methicillin resistant strains (MRSA), but community-onset infections (Co-MRSA) with fatal outcome in otherwise healthy young people were also reported in recent years [2]. In addition to acute infections, chronic infections by S. aureus pose a real challenge since the bacteria are difficult, often impossible, to eradicate, even with adequate and extended antibiotic treatments. Importantly, these prolonged treatments of persistent infections lead to the selection of strains resistant to many antibiotics.
S. aureus is continuously confronted with a sophisticated and very efficient innate immune system. Through its evolutionary association with its host, it has, however, developed a myriad of different defence strategies (table 1 and fig. 1). First, the pathogen can interfere with the signalling that allows the attraction of neutrophils or the activation of the complement. Once in contact with host defence mechanisms, the pathogen actively counteracts reactive oxygen species, blocks the action of antimicrobial peptides, or produces phenol soluble modulins (PSMs) to escape from the phagosome. To avoid killing by host defence cells, these bacteria also produce a variety of toxins, and finally, the pathogen produces coagulase to convert fibrinogen into fibrin to help wall off the pathogen from host defence. Here we describe how this skilled pathogen can rapidly adapt to a variety of different growth conditions and modulate its gene expression in the presence of these host defence mechanisms by changes in transcription, RNA processing and RNA decay.
Table 1 Factors used by Staphylococcus aureus to counter host defences.
|
Protein or gene
|
Function
|
Reference
|
| FLIPr |
Protein that inhibits leucocyte response mediated by activation of FPR-like protein 1. FPR is a high affinity receptor for N-formyl-met-leu-phe signalling tripeptide |
Prat et al., 2006 [29] |
| CHIPS |
Binds C5aR and the formyl peptide receptor FPR |
de Haas et al., 2004 [3] |
| Capsule |
Polysaccharide capsule prevents phagocytosis and adherence |
George et al., 2015 [5] |
| SCIN |
Staphylococcal complement inhibitor interacts with the C3 convertase, C4b2a and C3bBb |
Rooijakkers et al., 2005 [6] |
| Ecb |
Extracellular complement binding protein blocks C3 and C5 convertases |
Jongerius et al., 2012 [7] |
| Efb |
Extracellular fibrinogen binding protein, blocks complement and binding of neutrophils to fibrinogen, and platelet aggregation |
Jongerius et al., 2012 [7] |
| Protease V8 (SspA) |
Inhibition of complement pathways |
Jusko et al., 2014 [8] |
| Aureolysin (Aur) |
Inhibition of complement pathways |
Jusko et al., 2014 [8] |
| Staphopain (ScpA, SspB) |
Cystein protease cleaving CXCR2 chemokine receptor |
Jusko et al., 2014 [8]; Laarman et al., 2012 [4] |
| Protein A |
Interacts with Fc region of IgG |
Atkins et al., 2008 [9] |
| Sbi |
Interacts with Fc region of IgG |
Atkins et al., 2008 [9]; Zhang et al., 1998 [83] |
| Dismutases (SodA, SodM) |
Conversion of superoxide to hydrogen peroxide |
Das et al., 2008 [10] |
| Catalase (KatA) |
Conversion of hydrogen peroxide to water and oxygen |
Cosgrove et al., 2007 [11] |
| Staphyloxanthin |
Antioxidant carotenoid |
Clauditz et al., 2006 [12] |
| DNases |
Cleaves DNA in neutrophil extracellular traps, NETs |
Berends et al., 2010 [14] |
| Dlt operon |
Addition of D-alanyl esters to teichoic acids to protect against α-defensins |
Collins et al., 2002 [17] |
| Phenol soluble modulins (PSM) |
Small amphipathic α-helical peptides |
Queck et al., 2008 [47] |
| α-toxin, hla |
Pore forming toxin, lyses human leucocytes, epithelial and endothelial cells, platelets |
Seilie and Bubeck Wardenburg, 2017 [18] |
| Panton Valentine leucodicin (PVL) |
Pore forming bi-component leukocidin |
Seilie and Bubeck Wardenburg, 2017 [18] |
| γ-haemolysin (HlgAB, HlgCB), |
Pore forming bi-component leukocidin |
Seilie and Bubeck Wardenburg, 2017 [18] |
| LukED, LukAB |
Pore forming bi-component leukocidins |
Seilie and Bubeck Wardenburg, 2017 [18] |
| Coagulase |
Activates prothrombin to induce blood coagulation |
Friedrich et al., 2003 [21] |
| von Willebrand factor binding protein |
Activates prothrombin to induce blood coagulation |
Kroh et al., 2009 [22] |
| Staphylokinase |
Plasminogen activator to form the active protease plasmin |
Bokarewa et al., 2006 [84] |
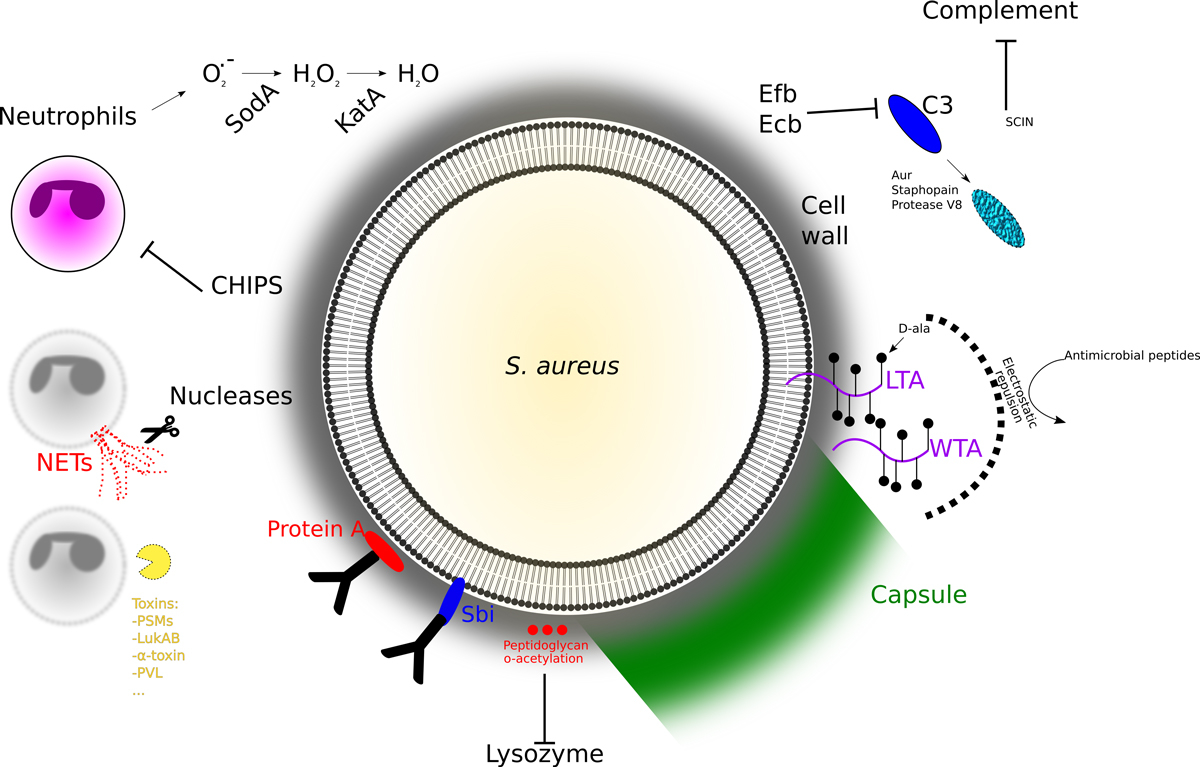
Figure 1
The battle between the pathogen and the host immune system. During infection, the bacteria encounter a series of efficient host defences that eliminate most pathogens. Although adaptative immunity certainly plays a role in our defence against S. aureus, it is mainly the ensemble of the actors of innate immunity that contain this opportunistic pathogen.
Interference with signalling for neutrophil recruitment and complement activation, weakens the host defence. Neutrophils are the first line of immune defence and circulate in the body to phagocyte and eliminate the pathogens. S. aureus secretes a variety of effectors that interfere with neutrophil recruitment, such as the CHIPS (chemotaxis inhibitory protein of S. aureus) [3], or staphopain (to cleave chemokine receptors) [4]. Another important host defence strategy is the activation of the complement with two main aims: decorating the bacteria with the opsonin C3b to facilitate opsonisation and to liberate the chemokine C5a to attract immune cells to site of infection. Decoration by opsonin is less efficient on capsulated bacteria, and some S. aureus strains produce capsules [5]. Interestingly, in a population capsular expression is heterogeneous and growth and medium dependent, multiplying the possibilities of the bacterium to face different environments. C3b production can also be inhibited by a small secreted peptide, SCIN, that binds to the C3 convertases (C4b2A and C3bBb), leading to a decrease of surface bound C3b [6]. Similarly, the extracellular complement binding protein Ecb and its homolog Efb (for extracellular fibrinogen binding protein) were shown to bind and inhibit C3 [7]. Moreover, many pathogenic bacteria including S. aureus, secrete proteases to inactivate complement proteins. For example, S. aureus secretes protease V8, aureolysin (Aur), and Staphopain A and B, interfering with the complement reaction [8]. Finally, S. aureus secretes cell-wall-bound or free proteins (protein A and the second immunoglobulin-binding protein, sbi) that bind immunoglobulins at their Fc region, thereby inhibiting the classical complement activation and phagocytosis [9].
The armour of the pathogen protects against host weapons. Although S. aureus has developed or acquired many different strategies to avoid phagocytosis, some cells will face a hostile environment if taken up by phagocytes. The phagocytic cell uses the NADPH oxidase (NOX) to produce superoxide (O2
-), which will be converted by dismutases to hydrogenperoxide (H2O2), which are bactericidal by creating hypochlorous acid using myeloperoxidase. To avoid killing by these harsh and bactericidal conditions, S. aureus has developed several ways to survive. It will accelerate the formation of H2O2 by superoxide dismutases [10], and then rapidly catalyse (catalase, KatA) the conversion of hydrogenperoxide into water and oxygen [11]. Finally, Staphyloxanthin, a powerful antioxidant responsible for the characteristic golden color of S. aureus also diminishes the damage by oxidative stress [12]. In some cases, the neutrophils sacrifice themselves to form the so-called NETs, Neutrophil Extracellular Traps [13]. This process, called NETosis, liberates the cellular chromatin trapping the bacteria in this network and killing them with the liberated content of the granules. The bacteria respond with the secretion of DNases to weaken the host defence [14]. Intriguingly, using this arsenal of virulence factors, S. aureus is even able to use the phagocytes as Trojan horses to disseminate [15, 16].
To avoid damage by granulosome contents, S. aureus modifies its peptidoglycan cell wall to make it resistant to lysozyme, insert molecules that inhibit binding of antimicrobial peptides (dlt operon for adding D-alanyl esters to teichoic acids) or degrade said peptides [17]. Trapped in a phagosome, S. aureus tries to escape into the cytoplasm by using phenol soluble modulines (PSMs) and other membrane damaging proteins.
In some cases counterattack is the best defence and the bacteria try to avoid the confrontation with host immune cells. S. aureus produces several toxins that lyse leukocytes. These toxins are divided into β-barrel toxins and bicomponent leukocidins. The 7-subunit α-toxin forms pores in platelets, endothelial cells, and leukocytes [18, 19]. The bicomponent toxins in human strains are composed of 8 subunits (Panton Valentine leukodicin (PVL), γ-haemolysin (HlgAB, HlgCB), LukED, LukAB). These toxins are not to be confounded with superantigens that activate non-specifically T-cells causing cytokine storms.
Finally, walling off the pathogen may also block access of leukocytes to the site of infection. In contrast to non-aureus Staphylococci (historically called the albus Staphylococci, nowadays coagulase-negative Staphylococci), S. aureus encodes a coagulase that was investigated by Chapman and colleagues [20]. Coagulase activates prothrombin by binding, but without the activating cleavage [21], to become (staphylo-)thrombin, which will convert fibrinogen into fibrin. The fibrin deposition will help to wall-off the pathogen and prevent access of leukocytes. In addition to coa, the coagulase, S. aureus encodes a second protein, the von Willebrand factor binding protein (vWbp) that can activate prothrombin in a similar way [22]. To observe a clear effect in a mice infection system, both genes need to be inactivated [23]. Although it may be advantageous to wall-off from the immune system, bacteria may also want to disseminate. In the case of S. aureus, the secreted staphylokinase is expressed at high cell density and forms plasmin from plasminogen. Plasmin is required to digest fibrin, and thereby allows bacteria to escape, but the role of the staphylokinase in pathogenicity is not well established.
Transcription
Gene expression depends on many different elements, from transcription to decay of RNA and proteins. The first step in gene expression is promoter recognition by the RNA polymerase. For this recognition, the core enzyme, constituted of five subunits (α2ββ′ω), associates with a sigma (σ) factor responsible for promoter recognition (fig. 2A). Whereas the housekeeping sigma A (σA) is responsible for the recognition of the majority of promoters, sigma B (σB) plays an important role in the stress response in many low guanine-cytosine Gram-positive bacteria, such as Bacillus, Listeria and Staphylococcus (for review see [24]). In S. aureus, microarrays have shown that σB influences the expression of 251 open reading frames (ORFs; 198 positively, 53 negatively) [25]. In a genome-wide transcription start site analysis, we have identified 121 sites preceded by a typical σB promoter sequence [26]. Amongst the genes regulated by σB we find many genes important for host-pathogen interaction. Moreover σB regulates positively the transcription factors SarA, SarS and ArlRS, that will further contribute to the wide effects of the σB regulon [25]. Another example of indirect regulation is the control of small regulatory RNAs (sRNA, see below), such as RsaA that represses the transcription factor MgrA [27] that controls indirectly clumping [28]. Recently it was also shown that RsaA controls the expression of SsaA-like proteins that are involved in peptidoglycan synthesis and control of the anti-inflammatory protein FLIPr (fig. 1) [29, 30]. Finally it has been shown that σB is important for chronic infections as a result of intracellular survival and that sigB mutants are readily cleared from the host and fail to form small colony variants, SCVs [31]. SCVs show a reduced metabolism and readily survive within eukaryotic cells.
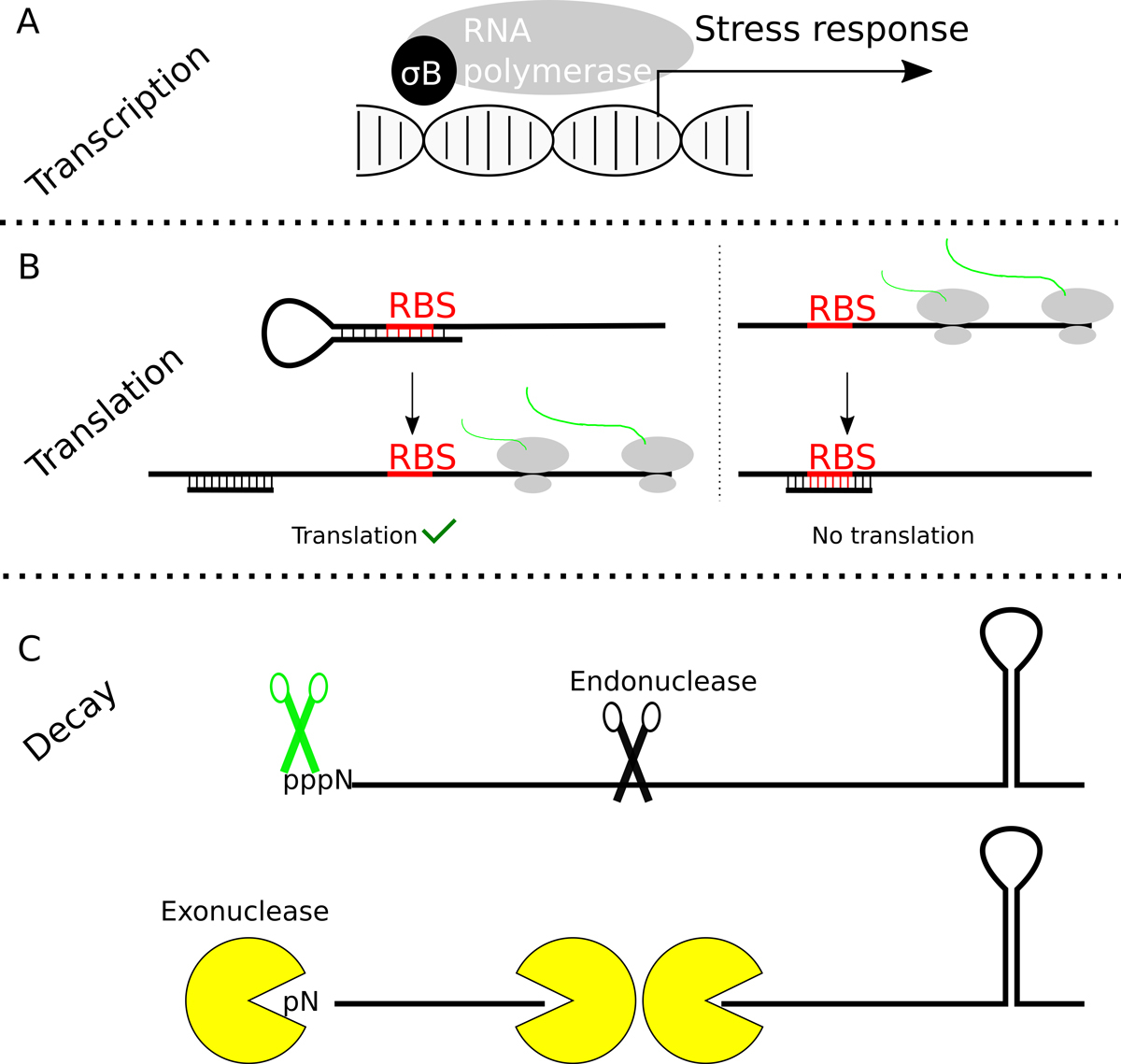
Figure 2
RNA synthesis, translation, processing and degradation. Gene expression can be regulated at many different levels. (A) Sigma factors direct the RNA polymerase to a subset of promoters and in the case of σB, the RNA polymerase will recognise promoters of stress response genes and operons. (B) To initiate translation, the small ribosomal subunit needs to recognise the RNA binding site by complementarity with its 16S rRNA. If the ribosome-binding site is trapped in a secondary structure or anneals with a sRNA, denaturation of the duplex will allow ribosome binding and translation (B, left). In some cases the RBS is normally accessible, but can be blocked by the synthesis and annealing of a sRNA (C, B, right). Several variations of these two possibilities are found in many bacteria. (C) To regulate gene expression the level of mRNAs may need to be carefully regulated. Dedicated enzymes, such endo – and exonucleases are involved in the processing and degradation of the RNAs. Additional enzymes, such as pyrophosphohydrolases (depicted as green scissors in the figure) or RNA helicases are required to render the RNA accessible for degradation, since triphosphorylated 5′ ends or secondary structures inhibit the exonucleases, respectively.
The sigma factor H has been shown to regulate several genes required for competence (DNA transformation) in S. aureus [32, 33]. S. aureus strains are known to encode many mobile elements, including bacteriophages, plasmids, chromosomal cassettes and pathogenicity islands, that are responsible for toxin production and the multiple antibiotic resistances these bacteria can acquire. Thus transformation, as a way of horizontal gene transfer, is very important. For a long time it was thought that S. aureus was not able to take up naked DNA by transformation, but Morikawa and co-worker showed that overexpression of σH leads to low frequency transformation. Using an elegant genetic screen it was later shown that σH expression can spontaneously occur upon genetic rearrangements in the S. aureus genome [32].
Thus, by selecting a subset of promoters, additional sigma factors can activate the transcription of genes required for the adaptation to particular growth conditions.
Translation
Once a messenger RNA (mRNA) is made, it is generally recognised at one or several ribosome binding sites (RBS), also called Shine-Dalgarno sequences, by the small ribosomal subunit. This recognition requires a complementary sequence in the 16S ribosomal RNA (rRNA) and the efficiency of translation depends on the quality of the hybridisation reaction. Thus, in contrast to eukaryotic translation, initiation in bacteria uses not a scanning mechanism, but rather internal binding sites. The control of translation on an individual mRNA can therefore be regulated by hiding the RBS, either by forming a secondary structure, by binding an antisense small RNA (sRNA), or by protein binding (fig. 2B). A magnificent example is the thermosensor in Listeria monocytogenes, where a secondary structure inhibits ribosome binding on the mRNA of the transcription factor PrfA [34]. At 37°C, however, the secondary structure is unfolded giving access to the ribosome and allowing translation. This in turn leads to the expression of virulence factors required during infection, but not during growth in the cold. To our knowledge, no thermosensor has yet been described in S. aureus, but they are present in other opportunistic pathogens inhabiting the nares, such as Neisseria meningitidis [35]. Nevertheless, an almost classical example for translational regulation by secondary structure is the pE194 plasmid-encoded combined resistance towards macrolides, lincosamides and streptogramin B through methylation of the ribosomal RNA [36] (fig. 3). This mRNA can form two secondary structures. A first structure can be made from repeats 1 and 2 or from repeats 2 and 3. This second possibility (2•3) is mutually exclusive with the second secondary structure, which is formed from repeats 3 and 4. Importantly, this second structure (3•4) blocks access of the ribosome for methylase translation. The clue in the regulation of translation initiation of the methylase is the presence of a short, 19 codon long, upstream ORF on the mRNA. If the ribosomes translating the leader peptide are inhibited by an antibiotic, such as erythromycin, half of the first secondary structure (repeat 1) is covered by ribosomes, thereby freeing repeat 2, which in turn will hybridise with repeat 3. The result of these interactions is that repeat 4 cannot be involved in secondary structure formation and the RBS for the methylase remains accessible, and the bacteria will methylate the rRNA and become antibiotic resistant [38]. In addition to this inducible antibiotic resistance, mutations can occur that have part of the regulatory region deleted and thereby constitutively express the methylase. In addition to this possibility to form intramolecular secondary structures, the S. aureus genome encodes many small non-coding RNAs that can anneal with mRNA and thereby influence translation efficiency, as described below.
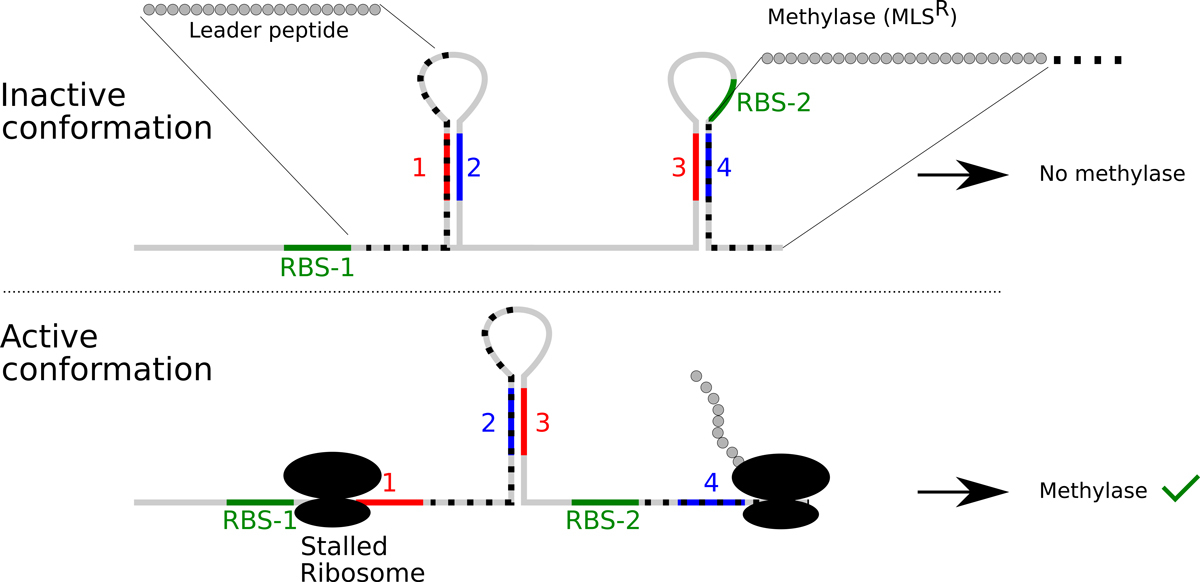
Figure 3
Regulation of macrolide-lincosamide-streptogramin B resistance. Resistance to these antibiotics occurs by methylating the ribosomal RNA to prevent binding of the antibiotic. The expression of the methylase is normally repressed by occluding the RBS in a secondary structure (3•4). In presence of the antibiotic, the translation of an upstream open reading frame will be blocked. This in turn will block the first inverted repeat and thereby allow the formation of the secondary structure 2•3. Since the inverted repeat 3 is already engaged, the inverted repeat 4 is free and the RBS for the methylase remains accessible, allowing expression of the resistance phenotype. Deletions can occur in the regulatory region and lead to constitutive MLS resistance [37].
Thus by modulating the access of the small ribosomal subunit to mRNA, the bacterium can very rapidly respond to environmental changes.
Small regulatory RNAs
Almost 40 years ago, the presence of a small antisense RNA was found to be involved in replication control of the Escherichia coli plasmid colEI [39, 40]. In this case, the RNA serving as primer for replication changes its conformation when hybridised with the sRNA, preventing RNase H cleavage of the RNA to serve as primer for replication initiation. Since then, the world of small regulatory RNAs has exploded, and nowadays many sRNAs are found to be involved in plasmid replication and virulence gene expression. In S. aureus, over 550 sRNAs are known and many of them regulate virulence factors [41]. These sRNAs can be cis-encoded, which means that they are produced from the complementary strand of the target mRNA and thereby fully match the target sequence. Others may be trans-encoded, i.e., they will act at one or more target genes located elsewhere on the genome. Through annealing with their target they can hide the ribosome binding site on mRNAs and thereby inhibit translation. In some cases, the annealing may also change the ability of mRNAs to fold into secondary structures and thereby allow or inhibit access of the ribosome to its binding site. In addition to an inhibitory function on translation initiation, double stranded RNA is a target for RNase III cleavage, which may initiate rapid RNA decay [42]. Finally, some sRNAs offer multiple protein binding sites, which will titrate out regulatory proteins that will no longer be available for their target RNAs [43, 44]. However such a regulatory mechanism has, to the best of our knowledge, not yet been described for S. aureus.
A paradigm of a small regulatory RNAs in S. aureus is RNAIII, an effector molecule of the agr quorum sensing system [45, 46]. The agrBDCA operon encodes a small peptide (AgrD or autoinducing peptide, AIP) that is secreted by AgrB. At high cell density, sufficient AIP is produced to successfully bind to the AgrC two-component receptor (fig. 4). Binding of AIP to AgrC induces autophosphorylation and phosphotransfer to AgrA, which then will be able to act as transcription factor. Activated AgrA induces transcription of the genes encoding phenol-soluble modulins (PSMs), which are small amphipathic α-helical peptides making pores in host cell membranes [47]. Moreover, AgrA also induces transcription of both the agrBDCA operon (thereby inducing a positive feedback loop) and RNAIII from a divergent promoter. This sRNA, which also encodes the δ-toxin (a PSM), negatively regulates surface proteins and some regulatory proteins by blocking the ribosome-binding site and by presenting a target for RNase III to induce degradation of the target RNA [48, 49]. In addition, RNAIII binds to mRNAs of secreted toxins to unfold secondary structures and thereby free the ribosome-binding sites, allowing translation of these proteins [50, 51].
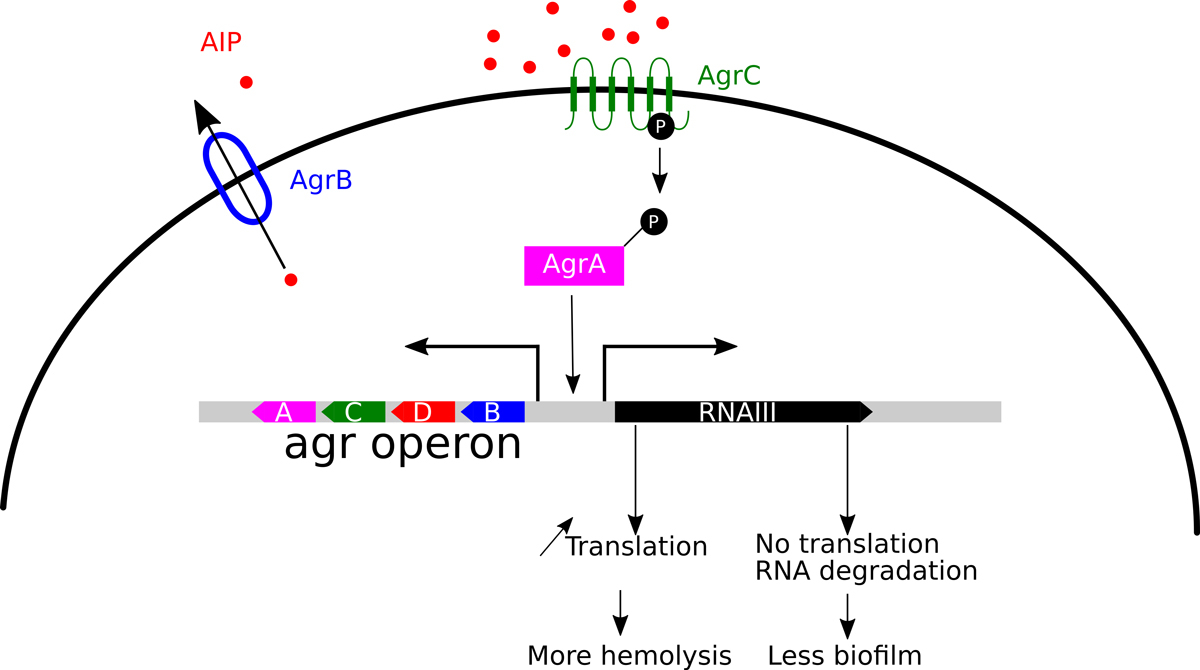
Figure 4
The quorum sensing system. The quorum sensing system agr (for accessory gene regulator) is encoded by an operon. The first open reading frame encodes the transporter AgrB, which is required for secretion and processing of the AgrD, the auto-inducing peptide, AIP. At sufficiently high cell concentration or in constraint environments, the concentration of AIP reaches a threshold and induces the two components system AgrCA, where AgrC activates AgrA. AgrA is a transcriptional activator required for the expression of PSMs and the effector molecule RNA III. RNA III is a paradigm of a regulatory RNA. It binds to the RBS on the mRNA of many genes and inhibits thereby translation. In some instances it binds to upstream sequences, helping to unwind secondary structures that occlude RBS elements and in these cases RNA III acts positively on translation.
An example of a cis-encoded sRNA is the regulatory element controlling plasmid replication in many S. aureus plasmids belonging to the pSK1 and pSK41 families [52, 53]. These plasmids are present in many clinical strains, and encode toxins and antibiotic resistance genes, such as β-lactamases. It has been postulated that the mRNA of the RepA protein, required for initiation of plasmid replication, has a long 5′ untranslated region that can form different and mutually exclusive secondary structures. In this model, in presence of a cis-acting sRNA (RNAI), the beginning of the repA mRNA will be double stranded with RNA I, and the remaining part of the 5′UTR is folded in a secondary structure that is limiting translation initiation, and thereby replication.
The synthesis of small regulatory RNA molecules is a rapid way to influence translation and the stability of mRNAs. In addition to rapid response to changing conditions, the synthesis of an sRNA is not as energy consuming as that of a complicated regulatory protein.
RNA processing and decay
To adapt to changing environmental conditions, such as in an infection, it is not only necessary to turn on certain virulence genes, but also important to stop expression of genes that may hinder the infection process. Therefore it is important that RNAs can also be efficiently degraded. Bacterial mRNAs are protected on their 5′ end by the triphosphate of the first nucleotide and at their 3′ end by secondary structures, which impede the degradation by 5′-3′ and 3′-5′ exoribonucleases, respectively. Thus degradation may be initiated by removal of a pyrophosphate at the 5′ end or by an internal cleavage (fig. 2C). The removal of the 5′ pyrophosphate occurs by enzymes of the Nudix family, such as RppH, and was first described for E. coli [54]. The S. aureus genome encodes several of these enzymes, and it has not been established to what extent they influence RNA turnover and virulence [55]. Once the 5′ end becomes accessible, RNase J, in S. aureus a tetramer of RNase J1 and RNase J2, rapidly degrades the unprotected RNA. This 5′-3′ RNase, which is not present in all bacteria, was first described in B. subtilis [56, 57], but is also present in S. aureus [58, 59]. The initiating endonucleolytic cleavage is performed by RNase E in E. coli and by RNase Y or by RNase J in B. subtilis. Whereas the inactivation of RNase Y in B. subtilis has a major impact, it seems to be less important for general RNA degradation in S. aureus [60, 61]. It has been shown that RNase J can also have an endonucleolytic activity [62, 63]. Both RNase J1/J2 and RNase Y were found to be present in the so-called degradosome, an RNA-degrading complex analogous to the eukaryotic exosome. As in its eukaryotic counterpart, the bacterial degradosome also contains an RNA helicase. In S. aureus this is the DEAD-box protein CshA [59, 64]. The bacterial degradosome also contain the metabolic enzyme enolase, which in some species can be replaced by aconitase and is supposed to link degradation to the metabolic activity of the cell [65].
One of the early indications that RNA turnover is important for virulence came from infection experiments in the silk worm [66]. In this experiment, 100 genes conserved amongst pathogenic bacteria were inactivated and the resulting bacteria injected into the haemolymph of the worms. From these, three candidates with attenuated virulence were further investigated (cvfA/SA1129 encoding RNase Y, cvfB/SA1223, cvfC/SA1262). An analysis of the agr mRNA showed decreased expression of the operon and its effector RNAIII in the RNase Y mutant. It was later shown that cvfA mutants are attenuated in a mice bacteraemia model using S. aureus Newman strains (see below) [67].
Both the RNase J genes and the gene for the RNA helicase CshA were identified in a screen for mutants deficient in biofilm formation [68]. We have subsequently shown that the inactivation of CshA results in increased stability of the agr mRNA and thereby in an increase of RNAIII [69]. It is likely that this increase of RNAIII results in reduced surface attachment, i.e., biofilm formation, and increased secretion of exoproteins, such as haemolysins (fig. 4).
Very recently, a beautiful example of mRNA processing by RNase Y and its influence on gene expression has been reported [70]. The SaePQRS operon (Sae for S. aureus exoprotein expression [71]) encodes the two-component system (TCS) SaeRS that is involved in the regulation of haemolysins, nucleases and coagulase upon recognition of signals from the human defence system, such as α-defensins or hydrogen peroxide [72]. The SaeP and SaeQ proteins are accessory proteins, whereas the SaeR and SaeS proteins constitute the TCS transcription factor and the sensor kinase, respectively. Marincola and Wolz showed that RNase Y is required for cleavage of the full-length transcript resulting in a stable downstream mRNA encoding the TCS components and a rapidly degraded upstream RNA fragment [67, 70]. It has been shown that mutations in the sae TCS strongly attenuate damage caused by the bacteria on macrophages through the induction of the Panton-Valentine leukocidin (PVL) and the LukAB leukocidin [73]. Thus, RNase Y, together with exoribonucleases, allows differential expression of genes involved in virulence regulation.
Thus, processing and RNA degradation are important to maintain cellular equilibria and to eventually rapidly change expression profiles.
Toxin-antitoxin systems
The toxin/antitoxin (TA) systems were originally discovered by studying partitioning systems in plasmids (for reviews see [74, 75]). The principle is the presence of an unstable antitoxin that inhibits the function of the toxin, but the toxin will become active when the antitoxin is no longer produced because the plasmid is lost or the antitoxin level is reduced for any other reason. These systems have also been termed addiction systems, since bacteria that lose a plasmid would die. Different types of TA systems are classified according to their molecular composition and function. In S. aureus three systems are known as of today [76]. The type I system consist of a membrane damaging peptide, the toxin, and an RNA that inhibits the translation of the peptide either by blocking translation or by inducing degradation of the toxin’s mRNA. The release of membrane-damaging toxin will arrest growth and kill the bacteria, but can also contribute to the escape of bacteria from vacuoles and damage competing bacteria or erythrocytes. In this case the toxin-producing bacteria would act like the Swiss Winkelried (who sacrificed himself in the battle of Sempach in 1386 to give victory to his confederates), sacrificing themselves for the survival of their sisters [77]. The type II systems are composed of two peptides, plasmid or chromosome encoded. The toxins of these systems are RNases that cleave mRNAs in a ribosome-dependent or -independent fashion, and thereby arrest growth of the bacteria [78]. The type III TA systems are composed of a toxin protein that is neutralised by binding to an RNA pseudoknot [79]. Per se, these systems are so far not known to participate in virulence expression. Nevertheless, the faithful maintenance of plasmids, bacteriophages or pathogenicity islands can contribute to the pathogenicity of the bacteria. Moreover, it was shown in E. coli that ciprofloxacin treatment induces a toxin/antitoxin system to block bacterial growth, and thereby cause persistent infections, despite the sensitivity of the bacterium to the antibiotic [80]. To our knowledge, such a situation has not (yet) been described for S. aureus.
Conclusion
Many pathogenic bacteria, including Staphylococcus aureus, face different environmental conditions to which they need to adapt rapidly. A common theme for these bacteria is the regulation of gene expression on the transcriptional and post-transcriptional level. In recent years, new-generation sequencing methods combined with more classical genetic tools have revealed a large diversity of regulatory levels. By these methods it has also become possible to monitor gene expression in infection systems, to better understand the behaviour of this feared pathogen. More recently, it was also proposed that RNA metabolism would be a suitable target in treating S. aureus infections [81, 82]. Although currently still a dream for the future, influencing RNA metabolism may become a way to eradicate dormant bacteria or other persistent infections.
Acknowledgements
We are grateful to J. Schrenzel and V. Khemici for careful reading of the present manuscript, and the reviewer for helpful suggestions to improve the text.
References
1
Wertheim
HF
,
Melles
DC
,
Vos
MC
,
van Leeuwen
W
,
van Belkum
A
,
Verbrugh
HA
, et al.
The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–62. doi:.https://doi.org/10.1016/S1473-3099(05)70295-4
2
Otto
M
. Community-associated MRSA: what makes them special?
Int J Med Microbiol. 2013;303(6-7):324–30. doi:.https://doi.org/10.1016/j.ijmm.2013.02.007
3
de Haas
CJ
,
Veldkamp
KE
,
Peschel
A
,
Weerkamp
F
,
Van Wamel
WJ
,
Heezius
EC
, et al.
Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J Exp Med. 2004;199(5):687–95. doi:.https://doi.org/10.1084/jem.20031636
4
Laarman
AJ
,
Mijnheer
G
,
Mootz
JM
,
van Rooijen
WJ
,
Ruyken
M
,
Malone
CL
, et al.
Staphylococcus aureus Staphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO J. 2012;31(17):3607–19. doi:.https://doi.org/10.1038/emboj.2012.212
5
George
SE
,
Nguyen
T
,
Geiger
T
,
Weidenmaier
C
,
Lee
JC
,
Liese
J
, et al.
Phenotypic heterogeneity and temporal expression of the capsular polysaccharide in Staphylococcus aureus. Mol Microbiol. 2015;98(6):1073–88. doi:.https://doi.org/10.1111/mmi.13174
6
Rooijakkers
SH
,
Ruyken
M
,
Roos
A
,
Daha
MR
,
Presanis
JS
,
Sim
RB
, et al.
Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol. 2005;6(9):920–7. doi:.https://doi.org/10.1038/ni1235
7
Jongerius
I
,
von Köckritz-Blickwede
M
,
Horsburgh
MJ
,
Ruyken
M
,
Nizet
V
,
Rooijakkers
SH
. Staphylococcus aureus virulence is enhanced by secreted factors that block innate immune defenses. J Innate Immun. 2012;4(3):301–11. doi:.https://doi.org/10.1159/000334604
8
Jusko
M
,
Potempa
J
,
Kantyka
T
,
Bielecka
E
,
Miller
HK
,
Kalinska
M
, et al.
Staphylococcal proteases aid in evasion of the human complement system. J Innate Immun. 2014;6(1):31–46. doi:.https://doi.org/10.1159/000351458
9
Atkins
KL
,
Burman
JD
,
Chamberlain
ES
,
Cooper
JE
,
Poutrel
B
,
Bagby
S
, et al.
S. aureus IgG-binding proteins SpA and Sbi: host specificity and mechanisms of immune complex formation. Mol Immunol. 2008;45(6):1600–11. doi:.https://doi.org/10.1016/j.molimm.2007.10.021
10
Das
D
,
Saha
SS
,
Bishayi
B
. Intracellular survival of Staphylococcus aureus: correlating production of catalase and superoxide dismutase with levels of inflammatory cytokines. Inflamm Res. 2008;57(7):340–9. doi:.https://doi.org/10.1007/s00011-007-7206-z
11
Cosgrove
K
,
Coutts
G
,
Jonsson
IM
,
Tarkowski
A
,
Kokai-Kun
JF
,
Mond
JJ
, et al.
Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. 2007;189(3):1025–35. doi:.https://doi.org/10.1128/JB.01524-06
12
Clauditz
A
,
Resch
A
,
Wieland
KP
,
Peschel
A
,
Götz
F
. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74(8):4950–3. doi:.https://doi.org/10.1128/IAI.00204-06
13
Brinkmann
V
,
Reichard
U
,
Goosmann
C
,
Fauler
B
,
Uhlemann
Y
,
Weiss
DS
, et al.
Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–5. doi:.https://doi.org/10.1126/science.1092385
14
Berends
ET
,
Horswill
AR
,
Haste
NM
,
Monestier
M
,
Nizet
V
,
von Köckritz-Blickwede
M
. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun. 2010;2(6):576–86. doi:.https://doi.org/10.1159/000319909
15
Thwaites
GE
,
Gant
V
. Are bloodstream leukocytes Trojan Horses for the metastasis of Staphylococcus aureus?
Nat Rev Microbiol. 2011;9(3):215–22. doi:.https://doi.org/10.1038/nrmicro2508
16
Voyich
JM
,
Braughton
KR
,
Sturdevant
DE
,
Whitney
AR
,
Saïd-Salim
B
,
Porcella
SF
, et al.
Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175(6):3907–19. doi:.https://doi.org/10.4049/jimmunol.175.6.3907
17
Collins
LV
,
Kristian
SA
,
Weidenmaier
C
,
Faigle
M
,
Van Kessel
KP
,
Van Strijp
JA
, et al.
Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis. 2002;186(2):214–9. doi:.https://doi.org/10.1086/341454
18
Seilie
ES
,
Bubeck Wardenburg
J
. Staphylococcus aureus pore-forming toxins: The interface of pathogen and host complexity. Semin Cell Dev Biol. 2017;S1084-9521(17)30207-0.
19
Spaan
AN
,
van Strijp
JAG
,
Torres
VJ
. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat Rev Microbiol. 2017;15(7):435–47. doi:.https://doi.org/10.1038/nrmicro.2017.27
20
Chapman
GH
,
Berens
C
,
Peters
A
,
Curcio
L
. Coagulase and Hemolysin Tests as Measures of the Pathogenicity of Staphylococci. J Bacteriol. 1934;28(4):343–63.
21
Friedrich
R
,
Panizzi
P
,
Fuentes-Prior
P
,
Richter
K
,
Verhamme
I
,
Anderson
PJ
, et al.
Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425(6957):535–9. doi:.https://doi.org/10.1038/nature01962
22
Kroh
HK
,
Panizzi
P
,
Bock
PE
. Von Willebrand factor-binding protein is a hysteretic conformational activator of prothrombin. Proc Natl Acad Sci USA. 2009;106(19):7786–91. doi:.https://doi.org/10.1073/pnas.0811750106
23
Cheng
AG
,
McAdow
M
,
Kim
HK
,
Bae
T
,
Missiakas
DM
,
Schneewind
O
. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 2010;6(8):e1001036. doi:.https://doi.org/10.1371/journal.ppat.1001036
24
van Schaik
W
,
Abee
T
. The role of sigmaB in the stress response of Gram-positive bacteria -- targets for food preservation and safety. Curr Opin Biotechnol. 2005;16(2):218–24. doi:.https://doi.org/10.1016/j.copbio.2005.01.008
25
Bischoff
M
,
Dunman
P
,
Kormanec
J
,
Macapagal
D
,
Murphy
E
,
Mounts
W
, et al.
Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol. 2004;186(13):4085–99. doi:.https://doi.org/10.1128/JB.186.13.4085-4099.2004
26
Prados
J
,
Linder
P
,
Redder
P
. TSS-EMOTE, a refined protocol for a more complete and less biased global mapping of transcription start sites in bacterial pathogens. BMC Genomics. 2016;17(1):849. doi:.https://doi.org/10.1186/s12864-016-3211-3
27
Romilly
C
,
Lays
C
,
Tomasini
A
,
Caldelari
I
,
Benito
Y
,
Hammann
P
, et al.
A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog. 2014;10(3):e1003979. doi:.https://doi.org/10.1371/journal.ppat.1003979
28
Crosby
HA
,
Schlievert
PM
,
Merriman
JA
,
King
JM
,
Salgado-Pabón
W
,
Horswill
AR
. The Staphylococcus aureus Global Regulator MgrA Modulates Clumping and Virulence by Controlling Surface Protein Expression. PLoS Pathog. 2016;12(5):e1005604. doi:.https://doi.org/10.1371/journal.ppat.1005604
29
Prat
C
,
Bestebroer
J
,
de Haas
CJ
,
van Strijp
JA
,
van Kessel
KP
. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J Immunol. 2006;177(11):8017–26. doi:.https://doi.org/10.4049/jimmunol.177.11.8017
30
Tomasini
A
,
Moreau
K
,
Chicher
J
,
Geissmann
T
,
Vandenesch
F
,
Romby
P
, et al.
The RNA targetome of Staphylococcus aureus non-coding RNA RsaA: impact on cell surface properties and defense mechanisms. Nucleic Acids Res. 2017;45(11):6746–60. doi:.https://doi.org/10.1093/nar/gkx219
31
Tuchscherr
L
,
Bischoff
M
,
Lattar
SM
,
Noto Llana
M
,
Pförtner
H
,
Niemann
S
, et al.
Sigma Factor SigB Is Crucial to Mediate Staphylococcus aureus Adaptation during Chronic Infections. PLoS Pathog. 2015;11(4):e1004870. doi:.https://doi.org/10.1371/journal.ppat.1004870
32
Morikawa
K
,
Inose
Y
,
Okamura
H
,
Maruyama
A
,
Hayashi
H
,
Takeyasu
K
, et al.
A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells. 2003;8(8):699–712. doi:.https://doi.org/10.1046/j.1365-2443.2003.00668.x
33
Morikawa
K
,
Takemura
AJ
,
Inose
Y
,
Tsai
M
,
Nguyen Thi
T
,
Ohta
T
, et al.
Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 2012;8(11):e1003003. doi:.https://doi.org/10.1371/journal.ppat.1003003
34
Johansson
J
,
Mandin
P
,
Renzoni
A
,
Chiaruttini
C
,
Springer
M
,
Cossart
P
. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110(5):551–61. doi:.https://doi.org/10.1016/S0092-8674(02)00905-4
35
Barnwal
RP
,
Loh
E
,
Godin
KS
,
Yip
J
,
Lavender
H
,
Tang
CM
, et al.
Structure and mechanism of a molecular rheostat, an RNA thermometer that modulates immune evasion by Neisseria meningitidis. Nucleic Acids Res. 2016;44(19):9426–37.
36
Mayford
M
,
Weisblum
B
. Messenger RNA from Staphylococcus aureus that specifies macrolide-lincosamide-streptogramin resistance. Demonstration of its conformations and of the leader peptide it encodes. J Mol Biol. 1985;185(4):769–80. doi:.https://doi.org/10.1016/0022-2836(85)90061-0
37
Catchpole
I
,
Dyke
KG
. A Staphylococcus aureus plasmid that specifies constitutive macrolide-lincosamide-streptogramin B resistance contains a novel deletion in the ermC attenuator. FEMS Microbiol Lett. 1990;69(1-2):43–7. doi:.https://doi.org/10.1111/j.1574-6968.1990.tb04172.x
38
Horinouchi
S
,
Weisblum
B
. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci USA. 1980;77(12):7079–83. doi:.https://doi.org/10.1073/pnas.77.12.7079
39
Tomizawa
J
. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984;38(3):861–70. doi:.https://doi.org/10.1016/0092-8674(84)90281-2
40
Tomizawa
JI
,
Itoh
T
. The importance of RNA secondary structure in CoIE1 primer formation. Cell. 1982;31(3 Pt 2):575–83. doi:.https://doi.org/10.1016/0092-8674(82)90313-0
41
Sassi
M
,
Augagneur
Y
,
Mauro
T
,
Ivain
L
,
Chabelskaya
S
,
Hallier
M
, et al.
SRD: a Staphylococcus regulatory RNA database. RNA. 2015;21(5):1005–17. doi:.https://doi.org/10.1261/rna.049346.114
42
Huntzinger
E
,
Boisset
S
,
Saveanu
C
,
Benito
Y
,
Geissmann
T
,
Namane
A
, et al.
Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24(4):824–35. doi:.https://doi.org/10.1038/sj.emboj.7600572
43
Liu
MY
,
Gui
G
,
Wei
B
,
Preston
JF, 3rd
,
Oakford
L
,
Yüksel
U
, et al.
The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272(28):17502–10. doi:.https://doi.org/10.1074/jbc.272.28.17502
44
Papenfort
K
,
Vanderpool
CK
. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev. 2015;39(3):362–78. doi:.https://doi.org/10.1093/femsre/fuv016
45
Novick
RP
. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48(6):1429–49. doi:.https://doi.org/10.1046/j.1365-2958.2003.03526.x
46
Novick
RP
,
Geisinger
E
. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42(1):541–64. doi:.https://doi.org/10.1146/annurev.genet.42.110807.091640
47
Queck
SY
,
Jameson-Lee
M
,
Villaruz
AE
,
Bach
TH
,
Khan
BA
,
Sturdevant
DE
, et al.
RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32(1):150–8. doi:.https://doi.org/10.1016/j.molcel.2008.08.005
48
Boisset
S
,
Geissmann
T
,
Huntzinger
E
,
Fechter
P
,
Bendridi
N
,
Possedko
M
, et al.
Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21(11):1353–66. doi:.https://doi.org/10.1101/gad.423507
49
Geisinger
E
,
Adhikari
RP
,
Jin
R
,
Ross
HF
,
Novick
RP
. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61(4):1038–48. doi:.https://doi.org/10.1111/j.1365-2958.2006.05292.x
50
Liu
Y
,
Mu
C
,
Ying
X
,
Li
W
,
Wu
N
,
Dong
J
, et al.
RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett. 2011;585(6):899–905. doi:.https://doi.org/10.1016/j.febslet.2011.02.021
51
Morfeldt
E
,
Taylor
D
,
von Gabain
A
,
Arvidson
S
. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14(18):4569–77.
52
Kwong
SM
,
Lim
R
,
Lebard
RJ
,
Skurray
RA
,
Firth
N
. Analysis of the pSK1 replicon, a prototype from the staphylococcal multiresistance plasmid family. Microbiology. 2008;154(Pt 10):3084–94. doi:.https://doi.org/10.1099/mic.0.2008/017418-0
53
Kwong
SM
,
Skurray
RA
,
Firth
N
. Staphylococcus aureus multiresistance plasmid pSK41: analysis of the replication region, initiator protein binding and antisense RNA regulation. Mol Microbiol. 2004;51(2):497–509. doi:.https://doi.org/10.1046/j.1365-2958.2003.03843.x
54
Deana
A
,
Celesnik
H
,
Belasco
JG
. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451(7176):355–8. doi:.https://doi.org/10.1038/nature06475
55
Bonnin
RA
,
Bouloc
P
. RNA Degradation in Staphylococcus aureus: Diversity of Ribonucleases and Their Impact. Int J Genomics. 2015;2015:395753. doi:.https://doi.org/10.1155/2015/395753
56
Even
S
,
Pellegrini
O
,
Zig
L
,
Labas
V
,
Vinh
J
,
Bréchemmier-Baey
D
, et al.
Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33(7):2141–52. doi:.https://doi.org/10.1093/nar/gki505
57
Mathy
N
,
Bénard
L
,
Pellegrini
O
,
Daou
R
,
Wen
T
,
Condon
C
. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129(4):681–92. doi:.https://doi.org/10.1016/j.cell.2007.02.051
58
Linder
P
,
Lemeille
S
,
Redder
P
. Transcriptome-wide analyses of 5′-ends in RNase J mutants of a gram-positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 2014;10(2):e1004207. doi:.https://doi.org/10.1371/journal.pgen.1004207
59
Roux
CM
,
DeMuth
JP
,
Dunman
PM
. Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J Bacteriol. 2011;193(19):5520–6. doi:.https://doi.org/10.1128/JB.05485-11
60
Figaro
S
,
Durand
S
,
Gilet
L
,
Cayet
N
,
Sachse
M
,
Condon
C
. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J Bacteriol. 2013;195(10):2340–8. doi:.https://doi.org/10.1128/JB.00164-13
61
Khemici
V
,
Prados
J
,
Linder
P
,
Redder
P
. Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation. PLoS Genet. 2015;11(10):e1005577. doi:. Correction in: PLoS Genet. 2016:12(9):e1006320. https://doi.org/10.1371/journal.pgen.1005577
62
Hausmann
S
,
Guimarães
VA
,
Garcin
D
,
Baumann
N
,
Linder
P
,
Redder
P
. Both exo- and endo-nucleolytic activities of RNase J1 from Staphylococcus aureus are manganese dependent and active on triphosphorylated 5′-ends. RNA Biol. 2017. [Epub ahead of print] doi:.https://doi.org/10.1080/15476286.2017.1300223
63
Mathy
N
,
Hébert
A
,
Mervelet
P
,
Bénard
L
,
Dorléans
A
,
Li de la Sierra-Gallay
I
, et al.
Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol. 2010;75(2):489–98. doi:.https://doi.org/10.1111/j.1365-2958.2009.07004.x
64
Giraud
C
,
Hausmann
S
,
Lemeille
S
,
Prados
J
,
Redder
P
,
Linder
P
. The C-terminal region of the RNA helicase CshA is required for the interaction with the degradosome and turnover of bulk RNA in the opportunistic pathogen Staphylococcus aureus
. RNA Biol. 2015;12(6):658–74. doi:.https://doi.org/10.1080/15476286.2015.1035505
65
Redder
P
,
Hausmann
S
,
Khemici
V
,
Yasrebi
H
,
Linder
P
. Bacterial versatility requires DEAD-box RNA helicases. FEMS Microbiol Rev. 2015;39(3):392–412. doi:.https://doi.org/10.1093/femsre/fuv011
66
Kaito
C
,
Kurokawa
K
,
Matsumoto
Y
,
Terao
Y
,
Kawabata
S
,
Hamada
S
, et al.
Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol Microbiol. 2005;56(4):934–44. doi:.https://doi.org/10.1111/j.1365-2958.2005.04596.x
67
Marincola
G
,
Schäfer
T
,
Behler
J
,
Bernhardt
J
,
Ohlsen
K
,
Goerke
C
, et al.
RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol. 2012;85(5):817–32. doi:.https://doi.org/10.1111/j.1365-2958.2012.08144.x
68
Tu Quoc
PH
,
Genevaux
P
,
Pajunen
M
,
Savilahti
H
,
Georgopoulos
C
,
Schrenzel
J
, et al.
Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus
. Infect Immun. 2007;75(3):1079–88. doi:.https://doi.org/10.1128/IAI.01143-06
69
Oun
S
,
Redder
P
,
Didier
JP
,
François
P
,
Corvaglia
AR
,
Buttazzoni
E
, et al.
The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol. 2013;10(1):157–65. doi:.https://doi.org/10.4161/rna.22899
70
Marincola
G
,
Wolz
C
. Downstream element determines RNase Y cleavage of the saePQRS operon in Staphylococcus aureus. Nucleic Acids Res. 2017;45(10):5980–94. doi:.https://doi.org/10.1093/nar/gkx296
71
Giraudo
AT
,
Calzolari
A
,
Cataldi
AA
,
Bogni
C
,
Nagel
R
. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett. 1999;177(1):15–22. doi:.https://doi.org/10.1111/j.1574-6968.1999.tb13707.x
72
Geiger
T
,
Goerke
C
,
Mainiero
M
,
Kraus
D
,
Wolz
C
. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol. 2008;190(10):3419–28. doi:.https://doi.org/10.1128/JB.01927-07
73
Münzenmayer
L
,
Geiger
T
,
Daiber
E
,
Schulte
B
,
Autenrieth
SE
,
Fraunholz
M
, et al.
Influence of Sae-regulated and Agr-regulated factors on the escape of Staphylococcus aureus from human macrophages. Cell Microbiol. 2016;18(8):1172–83. doi:.https://doi.org/10.1111/cmi.12577
74
Hall
AM
,
Gollan
B
,
Helaine
S
. Toxin-antitoxin systems: reversible toxicity. Curr Opin Microbiol. 2017;36:102–10. doi:.https://doi.org/10.1016/j.mib.2017.02.003
75
Lobato-Márquez
D
,
Díaz-Orejas
R
,
García-Del Portillo
F
. Toxin-antitoxins and bacterial virulence. FEMS Microbiol Rev. 2016;40(5):592–609. doi:.https://doi.org/10.1093/femsre/fuw022
76
Schuster
CF
,
Bertram
R
. Toxin-Antitoxin Systems of Staphylococcus aureus. Toxins (Basel). 2016;8(5):140. doi:.https://doi.org/10.3390/toxins8050140
77
Sayed
N
,
Nonin-Lecomte
S
,
Réty
S
,
Felden
B
. Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J Biol Chem. 2012;287(52):43454–63. doi:.https://doi.org/10.1074/jbc.M112.402693
78
Schuster
CF
,
Mechler
L
,
Nolle
N
,
Krismer
B
,
Zelder
ME
,
Götz
F
, et al.
The MazEF Toxin-Antitoxin System Alters the β-Lactam Susceptibility of Staphylococcus aureus. PLoS One. 2015;10(5):e0126118. doi:.https://doi.org/10.1371/journal.pone.0126118
79
Blower
TR
,
Short
FL
,
Rao
F
,
Mizuguchi
K
,
Pei
XY
,
Fineran
PC
, et al.
Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012;40(13):6158–73. doi:.https://doi.org/10.1093/nar/gks231
80
Dörr
T
,
Vulić
M
,
Lewis
K
. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8(2):e1000317. doi:.https://doi.org/10.1371/journal.pbio.1000317
81
Eidem
TM
,
Lounsbury
N
,
Emery
JF
,
Bulger
J
,
Smith
A
,
Abou-Gharbia
M
, et al.
Small-molecule inhibitors of Staphylococcus aureus RnpA-mediated RNA turnover and tRNA processing. Antimicrob Agents Chemother. 2015;59(4):2016–28. doi:.https://doi.org/10.1128/AAC.04352-14
82
Morrison
JM
,
Dunman
PM
. The modulation of Staphylococcus aureus mRNA turnover. Future Microbiol. 2011;6(10):1141–50. doi:.https://doi.org/10.2217/fmb.11.102
83
Zhang
et al., 1998
84
Bokarewa
MI
,
Jin
T
,
Tarkowski
A
. Staphylococcus aureus: Staphylokinase. Int J Biochem Cell Biol. 2006;38(4):504–9. doi:.https://doi.org/10.1016/j.biocel.2005.07.005



