Figure 1
Cell biological functions of ESCRTs (endosomal sorting complexes required for transport).
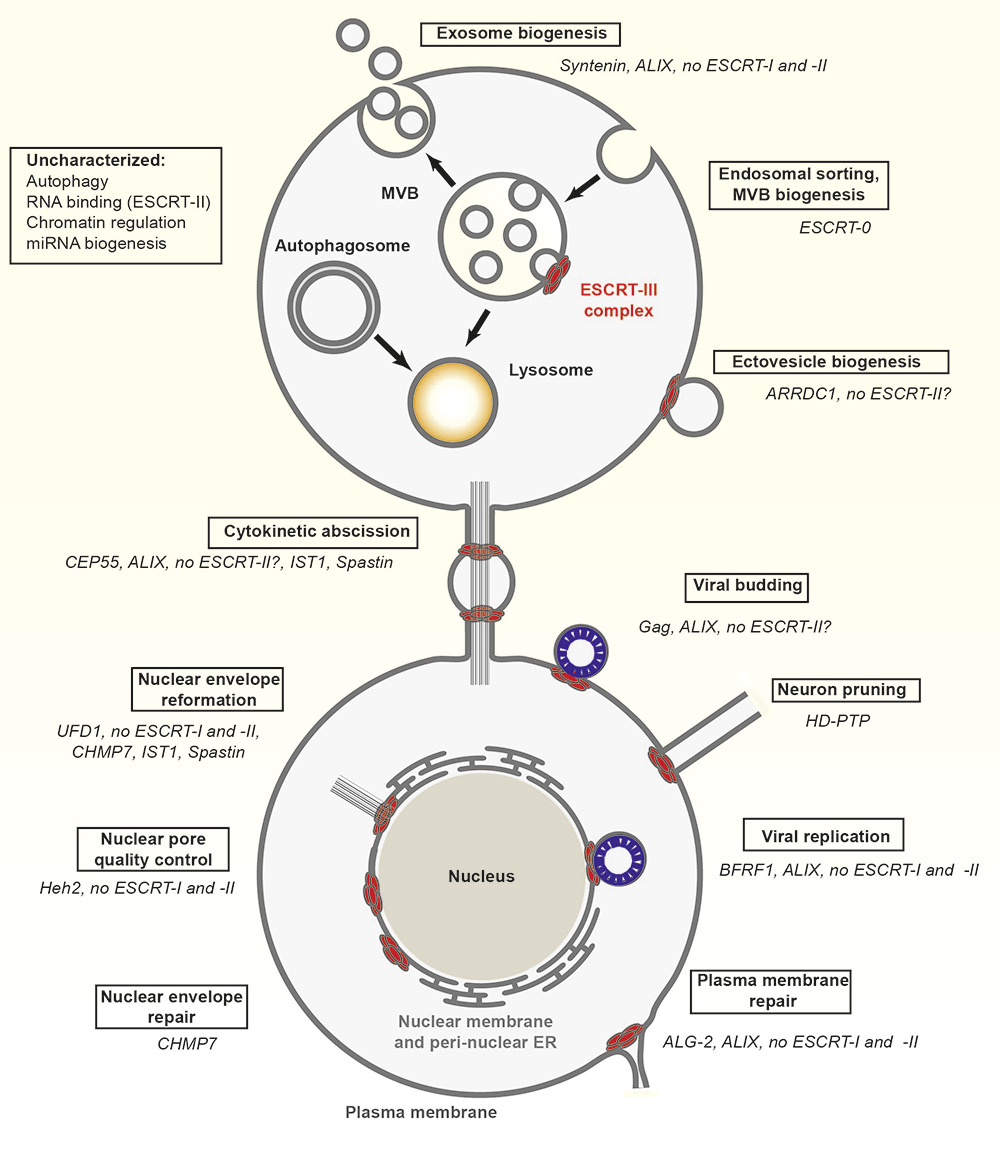
Overview of ESCRT functions throughout the cell. Factors that are specific for each ESCRT-dependent process (boxed) are listed in italics.
DOI: https://doi.org/10.4414/smw.2016.14347
Compartmentalisation is one of the defining features of eukaryotic cells. It is thought to have propelled life towards multi-cellularity and emergence of a nervous system [1, 2]. The plasma membrane and the endo-membranes of a compartmentalised cell constitute the infrastructure for most cellular logistics, which involves incessant trafficking of countless cargoes and associated macromolecules, ultimately shaping the identity and fate of a cell, as well as its relationship with neighbours. In this review, we focus on functions of the ESCRT (endosomal sorting complexes required for transport) machinery, which is emerging as a central regulator of membrane remodelling during trafficking and non-trafficking events (fig. 1).

Figure 1
Cell biological functions of ESCRTs (endosomal sorting complexes required for transport).
Overview of ESCRT functions throughout the cell. Factors that are specific for each ESCRT-dependent process (boxed) are listed in italics.
The ESCRT machinery was first identified in yeast by means of genetic isolation of mutants that cause defective protein sorting to the vacuole, the functional equivalent of the lysosome [3, 4]. These mutants, termed “class E-vps mutants”, possessed enlarged prevacuolar endosome-like compartments containing un-degraded proteins [5]. Most of the class E-vps genes were later found to act in succession to concentrate trafficking cargoes and include them in forming late endosomes (also termed multivesicular bodies or MVBs) that eventually fuse with lysosomes for degradation [6]. We now know from a large body of mechanistic studies in yeast and other model organisms that the ESCRT machinery that regulates endosomal sorting is organised into five distinct protein complexes: ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III and the Vps4 AAA-ATPase complex (Vps: vacuolar protein sorting-associated protein; AAA ATPases: ATPases Associated with diverse cellular Activities) (see table 1 for subunit compositions). During sorting, these complexes are recruited from the cytoplasm sequentially by interaction of specific subunits with the endosomal membrane. Ubiquitination of cargoes provides the key signal for initial cargo binding by ESCRT-0 (reviewed in [7]). Indeed, the ESCRT-0 subunits Hrs and Stam, as well as ESCRT-I Vps23/TSG101 and ESCRT-II Vps36, contain ubiquitin-binding domains that interact with ubiquitinated cargoes. ESCRT-0 is also recruited by interaction between the FYVE domain of Hrs and phosphatidylinositol 3-phosphate (PI3P), which is enriched at the endosomal membrane. ESCRT-0 is thought to concentrate ubiquitinated cargoes by organising flat coats of clathrin on the endosomal membrane [8, 9]. ESCRT-0 also summons ESCRT-I, which retains the cargoes by ubiquitin binding and hands them to the ESCRT–II complex. The ESCRT-II complex provides a scaffold for the formation of the ESCRT-III complex, the business end of the ESCRT machinery. The Vps32/Snf7/Chmp4 subunit of ESCRT-III forms multimeric filaments organised in spirals that bend the endosomal membrane away from the cytoplasm to form invaginated buds. Thus, the combined activity of ESCRTs allows sorted cargoes to be corralled and trapped in nascent intraluminal vesicles (ILVs) of the MVBs, which eventually pinch off into the endosomal lumen. The deubiquitinating enzyme Doa4 is recruited by ESCRT-III to remove ubiquitin from cargoes that are included into ILVs. Finally, the Vps4 ATPase complex binds and fully unfolds the ESCRT-III complex in an ATP-dependent manner and favours pinching off the ILV neck, the final step of MVB biogenesis [10–18]. The structure of most ESCRT components has been determined, and detailed extensive knowledge of the ESCRT mechanism of action in endosomal sorting and MVB biogenesis is available (reviewed in [19, 20])
| Table 1:Composition of the ESCRT (endosomal sorting complexes required for transport) complexes in endosomal sorting. | |||||
| Complex | Function | Evolutionary origin | Yeast | Drosophila | Human |
| ESCRT-0 | Cargo recognition | Opisthokonta | Vps27 | Hrs | HRS |
| Hse1 | Stam | STAM1, STAM2 | |||
| ESCRT-I | Upstream adapter | Eukaryotes | Vps23 | Tsg101 | TSG101 |
| Vps37 | VPS37A, B, C, D | ||||
| Mvb12 | Mvb12 | MVB12A, MVB12B | |||
| Vps28 | Vps28 | VPS28 | |||
| ESCRT-II | Bridging adapter | Vps36 | Vps36 | EAP45 | |
| Vs22(Snf8) | Vps22 | EAP30 | |||
| Vps25 | Vps25 | EAP20 | |||
| ESCRT-III | Membrane remodelling/filament | Archaea | Vps2 | Vps2 | CHMP2A, B |
| Vps24 | Vps24 | CHMP3 | |||
| Vps32 (Snf7) | Vps32 (Shrub) | CHMP4A, B, C | |||
| Vps20 | Vps20 | VPS20/CHMP6 | |||
| Vps46 (Did2) | Chmp1 | CHMP1A, B | |||
| Vps60 (Chm5) | Chmp5 | CHMP5 | |||
| Vps4 –Vta1 | Membrane remodelling/ATPase | Vps4 | Vps4 | VPS4A, B (SKD1, 2) | |
| Vta1 | CG7967 | VTA1 (LIP5) | |||
ESCRT-I, -II and -III and the Vps4 complexes are conserved across the eukaryotic lineage [21]. In contrast, ESCRT-0 is present only in a subset of eukaryotes. This indicated early on that they are specialised to couple the core membrane-remodelling activity of ESCRT-III and Vps4 with cargo sorting. Indeed, evidence indicates that additional complexes, such as those containing the protein Tom1 (target of Myb protein 1), might control initial concentration of ubiquitinated cargoes in endosomes [22]. Consistent with the accessory role of ESCRT-0, a large body of studies in the last 25 years revealed that the function of the ESCRTs at membranes is not limited to endosomal sorting and MVB biogenesis. In fact, early work indicated that a number of viruses can recruit ESCRT-III and Vps4 to bud from the plasma membrane [23, 24], leading to subsequent realisation that budding of the plasma membrane operated by ESCRTs occurs also in uninfected cells to form ectovesicles. MVBs can also fuse with the plasma membrane to release ILVs, in this case referred to as exosomes. As in endosomal sorting, deployment of ESCRT-III and Vps4 in the formation of exosomes and ectovesicles (together referred to as exovesicles) appears to depend either on ESCRT-I and -II, or on Alix, and to require adapters different from ESCRT-0. These data indicated that MVB and exovesicle biogenesis can profoundly differ and that multiple pathways of exovesicle formation are likely to exist [25–30].
MVBs also act as main stations for autophagic trafficking [31, 32]. Evidences from Caenorhabditis elegans, Drosophila and mammalian cells in culture revealed that ESCRTs are required for both microautophagy and macroautophagy [33–36]. During macroautophagy, autophagosomes that are formed de novo to clear long-lived proteins, cytoplasmic aggregates or damaged organelles fuse with MVBs and lysosomes to form amphisomes and autolysosomes, respectively, whose content is progressively degraded. Although it recently emerged that ESCRT activity is coordinated with macroautophagic response to starvation [37, 38], how ESCRT regulate autophagy mechanistically is currently unclear.
In summary, the membrane trafficking functions regulated by ESCRTs are crucial for lysosome-mediated cargo degradation, for release of exovesicles and, perhaps indirectly, for autophagy.
The first evidence of ESCRT functions that are independent of membrane trafficking indicated that ESCRT-III and Vps4 act at the plasma membrane to sever microtubules and release the midbody during cytokinesis. In this case, the recruitment is operated by the midbody protein Cep55 and ESCRT-III directly recruiting the microtubule-severing protein spastin. This activity is present also in Archaea and plants, suggesting that it is the most ancient evolutionarily [39–41]. Very recent studies showed similar recruitment of spastin during nuclear envelope reformation at the end of mitosis, albeit with a different recruitment system [42, 43]. However, the roles of ESCRTs at the nuclear membrane began to emerge with the recognition that ESCRTs are required for budding of the Epstein-Barr virus through the nuclear membrane [44]. More recently, it was found that the ESCRT machinery also restores membrane integrity after nuclear pore and nuclear envelope damage [43, 45, 46]. These membrane-repair functions of the ESCRT machinery are also observed at the plasma membrane [47] and a likely developmental counterpart of such activity has been observed in neuron remodelling. Indeed, ESCRTs have been shown to be required for the membrane scission that occurs in neuron pruning [48]. ESCRT-dependent neuronal remodelling events were described previously in Drosophila development, but had been attributed to endolysosomal trafficking of neuronal receptors [49–51].
Other, less understood, ESCRT functions include control of centrosome number during mitosis [52–55], transcriptional gene regulation [56–61], RNA transport [62], and microRNA biogenesis [63, 64]. Although the mechanistic details of these processes are unclear, it is reasonable to think that they might be linked to bending of membranes away from the cytoplasm, which to date represents the shared topological feature of well-characterised processes operated by ESCRT. In extreme synthesis, the ESCRT machinery is modular and invariantly leads to deployment of ESCRT-III and Vps4, a multipurpose membrane-remodelling complex.
Because a number of signalling proteins are transmembrane or membrane associated, endocytosis and trafficking to lysosomes are crucial to regulation of signal transduction (reviewed in [65]). Indeed, studies in cells of multicellular organisms that followed the initial discovery of ESCRT in yeast revealed that endosomal sorting complexes are essential to downregulate signalling, among which is that stimulated by epidermal growth factor (EGF) [66–74]. Subsequently, Drosophila mosaic animals also indicated that ESCRT function is required to regulate Notch signalling, which regulates multiple cell fate decisions. The canonical pathway is activated by binding of ligands to the transmembrane Notch receptor, triggering cleavage and translocation of an activated fragment, Notch intracellular domain, to the nucleus to de-repress transcription of target genes (reviewed in [75]). Drosophila organs developing in absence of ESCRT-I, -II or -III activity display increased, and for the most part ligand-independent, Notch signalling activity owing to accumulation on the limiting membrane of endosomes of Notch receptors that fail to be included into MVBs [76–80]. Despite endosomal Notch accumulation, ESCRT-0 mutant organs of mosaic animals do not show ectopic Notch signalling activity [67, 68, 81], highlighting differences in regulation of Notch signalling regulation by endosomal sorting, when compared with EGFR signalling.
In ESCRT-mutant Drosophila tissues, cell polarisation is lost, probably because a number of polarity determinants require endosomal trafficking to be maintained at correct levels to polarise membranes and cell-cell junctions [82–86]. Apoptotic response is enhanced as well. However, it is not clear whether this is an indirect consequence of the signalling and polarity defects [79, 87, 88]. In epithelial organs of Drosophila lacking ESCRT-I, -II or -III components, ectopic activation of signalling and altered cell polarity contribute to formation of tumour-like tissue that is highly over-proliferative, especially when apoptosis is inhibited. These traits led to the proposal that ESCRT genes act as tumour suppressors in metazoans (reviewed in [89]). Drosophila ESCRT-0 genes, however, do not behave as tumour suppressors, perhaps reflecting the distinct evolutionary origin of the complex [81].
Similarly to that of Drosophila, analysis of mouse ESCRT knock-outs revealed a requirement for cell survival, proliferation and signalling regulation leading to lethality early in embryogenesis [90–93]. Interestingly, a mouse hypomorph mutant of Vps25, encoding an ESCRT-II component, allows development to occur and reveals a specific requirement for ESCRTs in downregulating Sonic Hedgehog and fibroblast growth factor (FGF) signalling during limb development [94].
Several other signalling pathways have been shown to be deregulated when ESCRT function is impaired in multiple model systems. These include JNK (Jun amino-terminal kinases), JAK/STAT (Janus kinase / signal transducer and activator of transcription), Hedgehog, Wnt, FGF, Toll, nuclear factor kappa-beta (NFκβ) and transforming growth factor-beta (TGF-β) signalling [68, 76, 88, 93–101].
Overall, development and cell biology studies in multicellular organisms have taught us that ESCRTs have essential and pleiotropic functions that deeply impact tissue formation and homeostasis.
As introduced above, a number of pathogenic viruses including human immunodeficiency virus-1 (HIV-1), hepatitis C virus and Ebola virus hijack ESCRTs for their maturation and eventual budding to release infectious particles from infected cells (table 2). Indeed, plenty of data indicate that viral proteins, such as the Gag protein of HIV-1, recruit TSG101 and Alix, which in turn recruit ESCRT-III and Vps4 proteins, to the neck of the viral particle assembling at the plasma membrane [23, 102–108]. In the absence of TSG101 and ALIX, hepatitis C virus, herpes simplex virus type 1 (HSV-1), and to some extent, HIV-1 are still able to recruit ESCRT-III [102, 109, 110], suggesting that additional proteins mediate these interactions. Alternatively, viral proteins may be able to recruit downstream ESCRT components; for instance, the matrix protein VP40 of Ebola virus, in addition to recruiting TSG101, also directly recruits Vps4, with some other ESCRT proteins, to the site of budding [111].
In addition, a number of reports suggest that several viruses incorporate their proteins, messenger RNAs or microRNAs into exovesicles of their hosts to promote their spread, to modulate immunity, or to manipulate the microenvironment [112–119]. ESCRT activity is also required for entry of rotaviruses and human papilloma virus, as these are taken up by endocytosis, sorted into ILVs and eventually released in the cytoplasm [120–126]. Finally, a role for ESCRT-II in the replication of HIV-1 has also been reported. Depletion of ESCRT-II subunits in HIV-1-infected human HeLa cells affected the cytoplasmic trafficking of HIV-1 genomic RNA and reduced the expression of the HIV Gag protein [127, 128]. Similar results were reported for the hepatitis B virus [129]. Whether the function of ESCRT-II in this particular aspect of the viral life-cycle corresponds to that in transport of endogenous mRNA in Drosophila [62] is currently unclear.
Several non-viral pathogens also exploit the function of ESCRTs in infecting their hosts. Genome-wide screens in Drosophila S2 and murine macrophage cells have found that ESCRT components restrict the growth of mycobacteria by impairing phagosome maturation, raising the possibility that mycobacteria may disrupt host ESCRT function for their growth. Indeed, a protein secreted by Mycobacterium tuberculosis binds to Hrs to hinder sorting towards the lysosome for degradation [130–132]. A subunit of the lethal anthrax toxin secreted by Bacillus anthracis is packaged into ILVs of infected cells, both for a longer half-life and for exosomal secretion [133]. Finally, Candida albicans, an opportunistic fungal pathogen that colonises mucosal surfaces, requires ESCRT activity for pathogenesis and colonisation. In contrast to viruses and other pathogens that hijack the host ESCRT machinery, C. albicans uses its own ESCRT complex to adapt to the neutral–alkaline pH of the host environment [134–139].
In summary, viruses and other pathogens clearly exploit a wide range of the diverse cell biological functions of ESCRTs, offering multiple points of entry for future innovative therapies.
| Table 2:Involvement of ESCRT (endosomal sorting complexes required for transport) components in disease. | |||
| ESCRT complex | Subunit | Disease/dysfunction | References |
| Infections | |||
| ESCRT-0 | Hrs | Exosomal secretion of hepatitis C virus Mycobacterium tuberculosisresistance to degradation | [115] [132] |
| ESCRT-I/III/VPS4 | TSG101, CHMPs, VPS4 | Budding of viruses including HIV-1, Ebola. | Reviewed in [251] |
| ESCRT-II | EAP20, EAP45 | Replication of HIV-1 | [127, 128] |
| Cancer | |||
| ESCRT-0 | Hrs | Tumorigenesis and metastasis of HeLa cells | [166] |
| ESCRT-I | TSG101 | Cervical cancer | [141] |
| Breast cancer | [145, 150] | ||
| Lung cancer | [146] | ||
| Gallbladder adenocarcinoma | [148] | ||
| Ovarian carcinoma | [149, 150, 152] | ||
| VPS37A/HCRP1 | Hepatocellular carcinoma | [153, 154] | |
| Breast cancer | [155] | ||
| Ovarian cancer | [156] | ||
| Renal cell carcinoma | [157] | ||
| ESCRT-III | CHMP1A | Renal cell carcinoma | [158] |
| Pancreatic carcinoma | [159, 160] | ||
| CHMP4B | Hepatocellular carcinoma | [161] | |
| CHMP4C | Lung cancer | [162] | |
| VPS4 | VPS4A | Hepatocellular carcinoma | [163] |
| Ovarian carcinoma | [165] | ||
| Neurodegeneration | |||
| ESCRT-I | VPS37A | Hereditary spastic paraplegia | [182] |
| ESCRT-III | CHMP2B | Frontotemporal dementia | [167, 168] |
| Amyotrophic lateral sclerosis | [176, 178] | ||
| Alzheimer’s disease, dementia with Lewy bodies | [186, 188–190] | ||
| Other diseases | |||
| ESCRT-III | CHMP4B | Progressive childhood posterior subcapsular cataracts | [194] |
| VPS4 | VPS4 | Crohn’s disease | [196] |
Misexpression of ESCRT subunits has been associated with several types of human cancer. However, the role of ESCRT in tumorigenesis remains highly controversial. One of the most studied ESCRTs in this regard is the ESCRT-I gene TSG101, which was initially isolated in a search for novel tumour suppressor genes (TSG101: tumour susceptibility gene 101). Inactivation of TSG101 in NIH3T3 cells gave rise to metastatic tumours when xenografted in nude mice [140]. Consistent with this, TSG101 expression is significantly downregulated in cervical carcinomas [141]. Despite this, the role of TSG101 as a tumour suppressor has been debated, because it was later found that conditional knock-out of Tsg101 in mouse mammary epithelia did not promote tumour formation but arrested cell growth [142, 143]. Although TSG101 expression seems tightly regulated by an active mechanism [144], a study evaluating the effect of TSG101 overexpression indicated that tumour maintenance and progression rather than initiation might benefit from higher levels of TSG101 [145]. Despite this, the gene has been found to be significantly overexpressed also in lung cancer [146], gallbladder adenocarcinoma [147], papillary thyroid tumours [148] and ovarian carcinomas [149]. These tumours might be addicted to high levels of TSG101, as its depletion was shown to reduce tumour growth, to slow tumour migration, to halt cell cycle progression and to trigger apoptosis of cancer cells [150–152]. TSG101 appears also to be a prognostic marker in some cancers because its high expression correlates with poor prognosis, decreased survival, high tumour stage, and increased metastasis and invasion [147, 152].
Besides TSG101, another ESCRT-I gene, VPS37A, was identified because of its down-regulation in hepatocellular carcinomas and named human hepatocellular protein 1 (HCRP1) accordingly [72, 153]. Reduced VPS37A/HCRP1 expression strongly correlates with depth of tumour invasion, lower survival and higher rate of disease recurrence not only in hepatocellular carcinoma, but also in breast cancer, renal cell carcinoma, and oral and oropharyngeal cancers [153–157]. Most of the effect of VPS37A loss has been attributed to reduced EGF receptor degradation [72, 156], activation of downstream MAPK/ERK signalling and increased matrix metalloproteinase-2 (MMP2) expression: the loss of VPS37A has been suggested to increase tumour proliferation and invasion, and in ovarian cancer patients to lower response to cetuximab treatment [153, 156].
Several subunits of the human ESCRT-III and Vps4 complexes have been also linked to tumour development. CHMP1A appears significantly downregulated in renal cell carcinomas [158] and pancreatic tumours [159, 160], in which it has been suggested to function as a tumour suppressor. Accordingly, non-tumorigenic human embryonic kidney cells acquire the ability to form xenograft tumours when CHMP1A is depleted [160]. CHMP1A overexpression inhibits the proliferation of renal [158] and pancreatic tumour cells [160]. CHMP1A appears to inhibit tumour growth in the pancreas by regulating the activation of ataxia telangiectasia mutated (ATM) kinase and phosphorylation of p53 [159, 160]. Recent reports have identified a strong upregulation (and correlation with poor prognosis) of CHMP4B in hepatocellular carcinoma, and have suggested that CHMP4B and CHMP4C might be required to sustain proliferation and resistance to anticancer treatment in human hepatocellular and lung cancer cell lines, respectively [161, 162]. In a study aimed at characterising microRNAs in exosomes of hepatocellular carcinoma cells, it was found that modulation of Vps4A changed exosome content and activity. Vps4A was also found to act as a tumour suppressor, by repressing the PI3K/Akt pathway [163]. Other studies suggested that VPS4A and exosomes could influence resistance to cancer drugs like cisplatin and doxorubicin by modulating their efflux [164, 165].
Finally, expression of the ESCRT-0 component HRS is significantly increased in human tumour tissues derived from the stomach, colon, liver and cervix and from melanomas ― suggesting the existence of a tumour-enhancing function for HRS. Depletion of HRS reduced the tumorigenicity and metastatic ability of HeLa cells and upregulated the protein level of adherens junction component E-cadherin [166]. Since HRS functions in the endolysosomal trafficking and degradation of E-cadherin [83, 166], it has been proposed that, in these tumours, the cargo sorting function of HRS is hijacked to downregulate E-cadherin and promote metastasis.
Overall, the involvement of ESCRT in tumorigenesis is multifaceted and likely to be dependent on the tumour context, reflecting the complexity of the phenotypes observed in ESCRT mutant organs of Drosophila.
ESCRT loss is observed frequently in many neuropathologies. Among the best characterised are the form of autosomal dominant frontotemporal dementia (FTD) caused by mutations in CHMP2B, a subunit of ESCRT-III [167, 168]. The mutations lead to loss of the protein C terminus, which controls autoinhibition and interaction with Vps4 [169–173]. Accordingly, enlarged dysmorphic late endosomes have been found in cells of FTD patients [167, 174]. Similar endosomal phenotypes are observed when mutant CHMP2B is overexpressed in human cells [168]. It has been proposed that mutant CHMP2B impairs endosome-to-lysosome fusion by blocking the endosomal recruitment of the GTPase Rab7, by inhibiting ESCRT-III dissociation from endosomes, or by preventing the disassembly of the ESCRT-III complex. Defective autophagy is another mechanism by which CHMP2B mutations might cause FTD. Such a scenario is suggested by the presence of ubiquitin inclusions positive for the autophagy marker p62, which are often observed upon failure of autophagic clearance [173, 175–177]. Overall, the endolysosomal and autophagy defects are thought to lead to accumulation of protein aggregates, inducing neuronal degeneration, which is a hallmark of the disease. CHMP2B mutations have also been identified in amyotrophic lateral sclerosis patients [176, 178] suggesting that defective ESCRT activity may contribute also to the pathogenesis of amyotrophic lateral sclerosis.
Mutations in the microtubule-severing protein spastin, which has been found to be associated with the ESCRT-III complex during cytokinesis and nuclear membrane reformation, cause hereditary spastic paraplegia (HPS) [179]. Spastin function in HPS has been linked to shaping of the endoplasmic reticulum [180] and, recently, to formation of lipid droplets [181]. This indicates that either ESCRT-independent functions of spastin are affected in HPS or that ESCRTs and spastin might cooperate in membrane and microtubule remodelling at the endoplasmic reticulum or in lipid droplets. Underscoring this interesting possibility, mutations in VPS37 (ESCRT-I) have also been identified in HPS patients [182].
Although no mutations have been isolated so far, ESCRT-III function has also been reported to be important for aspects of Alzheimer’s disease and of Lewy body dementia (DLB, an umbrella term for two related diagnoses, Parkinson’s disease dementia and dementia with Lewy bodies). Lewy bodies are abnormal aggregates containing damaged alpha-synuclein (α-SYN) and other proteins, and α-SYN aggregation is a trait associated with the progression of Parkinson’s disease and DLB [183, 184]. A feature of Alzheimer’s disease and of DLB is the prion-like cell-to-cell spreading of α-SYN aggregates leading to rapid disease progression [185]. According to recent studies, α-SYN aggregates are taken up by clathrin-mediated endocytosis, undergo ESCRT-mediated trafficking through MVBs, and are degraded in lysosomes [186–188]. Rapid clearance of α-SYN aggregates and amelioration of the neurodegenerative pathology was observed upon CHMP2B overexpression [186, 189]; on the other hand, depletion of CHMP2B mediated by small interfering RNAs (siRNA) increased the exocytosis and intercellular transmission of α-SYN aggregates [186]. In addition, α-SYN aggregates colocalised with Vps4 [190], and inhibition of Vps4 function using a dominant-negative construct blocked lysosome-mediated degradation and increased extracellular secretion of α-SYN, possibly by means of exosomes [187].
The formation of amyloid-beta aggregates in Alzheimer’s disease also appears to involve regulation by ESCRT proteins. In fact, it has been recently shown that amyloid-beta and amyloid protein precursor are sorted into the intraluminal vesicles of MVBs. Depletion of Hrs and Tsg101 increases the intracellular accumulation of amyloid-beta by simultaneously inhibiting lysosomal delivery of amyloid precursor protein and reduced amyloid-beta secretion through an as yet unknown mechanism [191].
Finally, early work showed that fluorescently-tagged polyglutamine aggregates of mutant huntingtin protein required the function of the ESCRT-III protein CHMP3/Vps24 for autophagic clearance [192, 193]. However, no follow-up has further detailed alterations of ESCRT activity in Huntington’s disease.
Overall these studies clearly suggest that defects in endosomal sorting, autophagy, exosome release and spastin-dependent membrane remodelling contribute to key aspects of the pathology of a broad range of neurodegenerative diseases and that future modulation of ESCRT activity could provide a major therapeutic benefit.
Mutations in the ESCRT-III subunit CHMP4B have been identified in progressive childhood posterior subcapsular cataracts linked to chromosome 20q [194]. According to Sagona and colleagues, CHMP4B may protect the lens from developing cataract by mediating the autophagolysosomal degradation of micronuclei during lens differentiation, or by ensuring efficient cytokinesis [195]. Intestinal epithelial cells of patients with Crohn’s Disease, an inflammatory bowel disease, possess significantly upregulated Vps4B expression. This upregulation facilitates apoptosis of intestinal epithelial cells by activating the MAPK signalling pathway [196].
In recent years, we have witnessed a dramatic expansion in our knowledge of ESCRT activities. In fact, the current landscape of ESCRT-dependent processes covers a large palette of cellular events involving membrane remodelling, well beyond endosomal sorting. More are likely to surface in the next few years. Some of these are likely to explain the currently unclear involvement of ESCRTs in centrosome, chromatin and RNA regulation. However, it is already clear from the wealth of ESCRT functions, that in the future it will be critical to understand more about the factors and the modifications that regulate ESCRT activity in each different process. In this regard, we know that:
1. ESCRT targeting factors greatly differ. ESCRT-0 is used only for endosomal sorting, Gag for HIV-1 budding, CEP55 is specific for cytokinesis, syntenin and ARRDC1 for exo-vesicle secretion, BFRF1, Heh2 and UFD1 for ESCRT activities at the nuclear envelope and ALG-2 for plasma membrane wound repair.
2. ESCRT‐III and Vps4 can use ALIX as an alternative upstream ESCRT in HIV-1 release, cytokinesis, exosome formation, plasma membrane repair and Epstein-Barr virus budding from the nuclear envelope.
3. ESCRT-II appears dispensable as a bridging ESCRT during exosome biogenesis, plasma membrane repair and functions at the nuclear envelope.
4. Special ESCRT-III subunits, such as IST1, that assist cytokinesis and nuclear envelope reformation, or CHMP7 (also involved in nuclear envelope reformation) are often used.
5. A number of other components have been identified as accessory ESCRT-III subunits or regulators of the enzymatic activity of VPS4 with unclear specificity. They include Vta1/LIP5, Vps60/CHMP5, Did2/CHMP1 (reviewed in [197]).
Overall, these differences provide us with an initial glimpse of the functional diversity of ESCRT operations that will allow therapeutic targeting of distinct ESCRT processes in the future.
Our knowledge of the ESCRT modifications that might contribute to specificity and to functional modulation is unfortunately less developed. However, pioneering work has been done on post-translational modification of ESCRTs in the context of endosomal sorting. For instance, it has been shown early on that Hrs becomes rapidly phosphorylated in response to growth factors like HGF (hepatocyte growth factor), EGF and platelet-derived growth factor (PDGF) [198, 199] and subsequently ubiquitinated by the E3 ligase Cbl. Hrs modifications disrupt the interaction of its ubiquitin identification motif (UIM) domain with ubiquitinated (Ub-) cargoes and relocate Hrs to the cytosol to facilitate transfer of Ub-cargoes to downstream Ub-binding ESCRT proteins. Also, relocation of ubiquitinated Hrs from endosomal membranes permits replacement by non-ubiquitinated Hrs to sustain endosomal sorting [200–204]. At the level of ESCRT-I, mahogunin-1 monoubiquitinates TSG101 to favour endolysosomal cargo degradation [205–207], while TAL (Tsg101 associated ligase) specifically polyubiquitinates and aims Tsg101 towards degradation, thus inhibiting endosomal trafficking [208, 209]. Since evidence points to oncogenic alteration of signalling in tumours, and to toxic build-up of cargoes in neurodegeneration, such modifications provide potential targets for therapy.
The exact set of cargoes that can initiate modifications and which other cargo-specific factors can directly regulate ESCRT activity remains unresolved. Interesting in this context is the case of members of the Lgd/CC2D1 protein family (Lgd in Drosophila, CC2D1A and CC2D1B in mammals). They interact with CHMP4/Vps32 and appear required for the function of the CHMP4/Vps32 subunit of ESCRT-III complex. However, at least in Drosophila, Lgd regulation of ESCRT-III function appears highly specific to a limited subset of cargoes, as it leads only to alteration of Notch and BMP signalling [210–217]. Such example reminds us also of how little is still our understanding of how major signalling pathways are regulated by the ESCRTs. A case in point is that of Notch signalling, which is perturbed in a wide variety of cancers [218]. In Drosophila, Notch peculiarly appears to require fusion of MVBs to lysosomes to be ectopically activated by ESCRT impairment [219]. It is not yet clear whether such a form of regulation applies also to mammalian or cancer cells. However, pharmacological inhibition of endosomal acidification appears to reverse excessive Notch signalling also in mammalian cells [220], highlighting how complex and cargo-specific ESCRT regulation of signalling could be in health as well as disease.
In neuropathologies in which ESCRT mutations clearly lead to cargo accumulation, modifications and factors that might generally upregulate cellular clearance might have therapeutic benefits. A favourable strategy to achieve such a goal might be to activate the transcription factor EB (TFEB), a key regulator of lysosome biogenesis. TFEB activity drives autophagosome-lysosome fusion [221]. Phosphorylation of TFEB by the mechanistic target of rapamycin (mTOR) retains TFEB in the cytosol and prevents its translocation to the nucleus as part of the genetic circuitry that controls amino acid metabolism. Pharmacological inhibition of mTOR (e.g., by rapamycin, Torin1 or 2-hydroxypropyl-β-cyclodextrin) causes activation of TFEB [222–225] and together with TFEB overexpression has shown promise in models of neurodegenerative diseases characterised by autophagic accumulation of toxic protein [226–228]. TFEB overexpression appears to alter also Notch-related development signalling events in Drosophila [229], suggesting that the strategy might also beneficially modulate signalling originating from late endosomes.
A more complex example of ESCRT modification is that of Myopic (Mop, HD-PTP in mammals), a member of the protein tyrosine phosphatase (PTP) family that co-localises with ESCRT-0 on endosomes and promotes receptor trafficking towards the lysosome [96, 230–234]. Since the phosphatase activity of Mop is not important for this function and its human orthologue HD-PTP does not possess phosphatase activity [235, 236], exactly how Mop regulates the activity of ESCRT-0 is not well understood. It has been proposed that the PTP domain of HD-PTP/Mop might act as a phospho-tyrosine binding module preventing dephosphorylation or influencing the localisation of ESCRT-0 subunits [236]. Interestingly, HD-PTP activity is also required in lieu of ESCRT-II at the plasma membrane for neuron pruning [48], suggesting that interaction between ESCRTs and Mop might occur at multiple levels of membrane remodelling.
Modifications that specifically affect virus budding have also emerged recently. The primary defence against invading pathogens, including viruses, is the production of type-I interferon (IFN); pathogenic viruses have, however, evolved mechanisms to circumvent this control [237]. IFN upregulates the production of IFN-stimulated genes (ISGs). One of these ISGs is the ubiquitin-like ISG15, which has broad-spectrum antiviral activities [238–240]. Overexpression of ISG15 in HIV-1-infected human cells inhibited the replication of HIV-1 and disrupted the interaction of HIV-1 Gag protein with TSG101 by preventing Gag ubiquitination. [241]. This Gag-TSG101 interaction is crucial for efficient budding of HIV-1. Another mechanism of inhibition of HIV-1 budding by ISG15 is the disruption of VPS4 interaction with LIP5/Vta1 by ISGylating CHMP5 (ISGylation is a ubiquitin-like modification). In the absence of LIP5/Vta1, VPS4 fails to oligomerise and is retained in the cytoplasm. This results in failure to disassemble the ESCRT-III complex required for multiple rounds of scission, thus blocking virus budding [242–245]. The effect of ISG15 on host endosomal sorting has not been fully studied, so it is difficult to predict the level of cytotoxicity associated with this therapy. Gag-TSG101 interaction is also specifically inhibited by overexpression of the N-terminal Gag-binding domain of TSG101 (TSG-5’). Because host endosomal sorting remains relatively intact after TSG101-5’ overexpression, some authors proposed the development of TSG101-5’ derivatives or similarly-acting Gag-TSG101 inhibitors as specific and potent antiviral therapies [246–248]. Inhibiting the interaction of Ebola virus VP40 protein with ESCRT subunits has also been proposed as a therapeutic strategy [249]. Based on all these observations, small molecules have been, and are still being, developed to disrupt the interaction of viral proteins with host ESCRT or ESCRT-associated proteins [250]. A greater understanding of the cofactors that mediate ESCRT-II function in HIV-1 replication will also aid in the development of appropriate inhibitors at this step. Finally, understanding if these therapeutic strategies and the underlying mechanisms of actions are applicable to other types of ESCRT-mediated viral infections will be of great benefit to public health.
In conclusion, the recent explosion of studies documenting the new functions of ESCRT in membrane remodelling and the increasing evidence of deregulation of ESCRT in a wide range of pathologies demand that we step up our effort towards a deeper and process-specific understanding of ESCRT function, both in physiology and in disease. Such understanding will be invaluable for future preventive medicine and disease treatment.
Acknowledgements: We apologize to the colleagues whose work has not been referenced here due to space constraints. Work in the T. V. lab is supported by AIRC Investigator Grant #15954, by the Telethon Grant GPP13225 and by the Cariplo Foundation.
1 Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354.
2 Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci. 2007;120:2977–85.
3 Rothman JH, Howald I, Stevens TH. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989;8:2057–65.
4 Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci U S A. 1986;83:9075–9.
5 Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–402.
6 Katzmann DJ, Babst M, Emr SD. Ubiquitin-Dependent Sorting into the Multivesicular Body Pathway Requires the Function of a Conserved Endosomal Protein Sorting Complex, ESCRT-I. Cell. 2001;106:145–55.
7 Urbé S. Ubiquitin and endocytic protein sorting. Essays Biochem. 2005;41:81–98.
8 Raiborg C, Bache KG, Mehlum A, Stang E, Stenmark H. Hrs recruits clathrin to early endosomes. EMBO J. 2001;20:5008–21.
9 Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–8.
10 Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–9.
11 Malerød L, Stenmark H. ESCRTing membrane deformation. Cell. 2009;136:15–7.
12 Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–83.
13 Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne J a, Katzmann DJ, Piper R. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol. 2009;185:213–24.
14 Mageswaran SK, Dixon MG, Curtiss M, Keener JP, Babst M. Binding to any ESCRT can mediate ubiquitin-independent cargo sorting. Traffic. 2014;15:212–29.
15 Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162:435–42.
16 Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–71.
17 Adell MAY, Vogel GF, Pakdel M, Muller M, Lindner H, Hess MW, Teis D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205:33–49.
18 Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat Struct Mol Biol. 2015;22:492–8.
19 Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–68.
20 Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–87.
21 Wideman JG, Leung KF, Field MC, Dacks JB. The cell biology of the endocytic system from an evolutionary perspective. Cold Spring Harb Perspect Biol. 2014;6:a016998.
22 Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic. 2009;10:161–71.
23 Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65.
24 Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–7.
25 Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–9.
26 Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012;109:4146–51.
27 Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–85.
28 Choudhuri K, Llodrá J, Roth EW, Tsai J, Gordo S, Wucherpfennig KW, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507:118–23.
29 MacDonald C, Payne JA, Aboian M, Smith W, Katzmann DJ, Piper RC. A family of tetraspans organizes cargo for sorting into multivesicular bodies. Dev Cell. 2015;33:328–42.
30 Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–65.
31 Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–72.
32 Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–74.
33 Rusten TE, Vaccari T, Lindmo K, Rodahl LMW, Nezis IP, Sem-Jacobsen C, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–25.
34 Roudier N, Lefebvre C, Legouis R. CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic. 2005;6:695–705.
35 Morelli E, Ginefra P, Mastrodonato V, Beznoussenko G V, Rusten TE, Bilder D, et al. Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy. 2014;10:2251–68.
36 Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–9.
37 Jones CB, Ott EM, Keener JM, Curtiss M, Sandrin V, Babst M. Regulation of membrane protein degradation by starvation-response pathways. Traffic. 2012;13:468–82.
38 Müller M, Schmidt O, Angelova M, Faserl K, Weys S, Kremser L, et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. Elife. 2015;4:e07736.
39 Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–12.
40 Lindås A-C, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A. 2008;105:18942–6.
41 Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17:507–13.
42 Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522:231–5.
43 Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–9.
44 Lee C-P, Liu P-T, Kung H-N, Su M-T, Chua H-H, Chang Y-H, et al. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus. PLoS Pathog. 2012;8:e1002904.
45 Webster BM, Colombi P, Jäger J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159:388–401.
46 Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362.
47 Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136.
48 Loncle N, Agromayor M, Martin-Serrano J, Williams DW. An ESCRT module is required for neuron pruning. Sci Rep. 2015;5:8461.
49 Sweeney NT, Brenman JE, Jan YN, Gao F-B. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr Biol. 2006;16:1006–11.
50 Zhang H, Wang Y, Wong JJL, Lim K-L, Liou Y-C, Wang H, Yu F. Endocytic pathways downregulate the L1-type cell adhesion molecule neuroglian to promote dendrite pruning in Drosophila. Dev Cell 2014;30:463–78.
51 Issman-Zecharya N, Schuldiner O. The PI3K class III complex promotes axon pruning by downregulating a Ptc-derived signal via endosome-lysosomal degradation. Dev Cell. 2014;31:461–73.
52 Frost A, Elgort MG, Brandman O, Ives C, Collins SR, Miller-Vedam L, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149:1339–52.
53 Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WIHuman ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A. 2010;107:12889–94.
54 Jin Y, Mancuso JJ, Uzawa S, Cronembold D, Cande WZ. The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev Cell. 2005;9:63–73.
55 Xie W, Li L, Cohen SN. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc Natl Acad Sci U S A. 1998;95:1595–600.
56 Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114:2383–93.
57 Kamura T, Burian D, Khalili H, Schmidt SL, Sato S, Liu WJ, et al. Cloning and characterization of ELL-associated proteins EAP45 and EAP20. a role for yeast EAP-like proteins in regulation of gene expression by glucose. J Biol Chem. 2001;276:16528–33.
58 Schmidt AE, Miller T, Schmidt SL, Shiekhattar R, Shilatifard A. Cloning and characterization of the EAP30 subunit of the ELL complex that confers derepression of transcription by RNA polymerase II. J Biol Chem. 1999;274:21981–5.
59 Burgdorf S, Leister P, Scheidtmann KH. TSG101 interacts with apoptosis-antagonizing transcription factor and enhances androgen receptor-mediated transcription by promoting its monoubiquitination. J Biol Chem. 2004;279:17524–34.
60 Sun Z, Pan J, Hope WX, Cohen SN, Balk SP. Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300. Cancer. 1999;86:689–96.
61 Lin Y-S, Chen Y-J, Cohen SN, Cheng T-H. Identification of TSG101 functional domains and p21 loci required for TSG101-mediated p21 gene regulation. PLoS One. 2013;8:e79674.
62 Irion U, St Johnston D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445:554–8.
63 Lee Y, Pressman S, Andress A, Kim K. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–6.
64 Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–9.
65 Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP. Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol Rev. 2012;92:273–366.
66 Chanut-Delalande H, Jung AC, Baer MM, Lin L, Payre F, Affolter M. The Hrs/Stam complex acts as a positive and negative regulator of RTK signaling during Drosophila development. PLoS One. 2010;5:e10245.
67 Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ. Hrs Regulates Endosome Membrane Invagination and Tyrosine Kinase Receptor Signaling in Drosophila. Cell. 2002;108:261–9.
68 Jékely G, Rørth P. Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 2003;4:1163–8.
69 Bache KG, Stuffers S, Malerød L, Slagsvold T, Raiborg C, Lechardeur D, et al. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol Biol Cell. 2006;17:2513–23.
70 Bishop N, Horman A, Woodman P. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J Cell Biol. 2002;157:91–101.
71 Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–58.
72 Bache KG, Slagsvold T, Cabezas A, Rosendal KR, Raiborg C, Stenmark H. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol Biol Cell. 2004;15: 4337–46.
73 Doyotte A, Russell MRG, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian “Class E” compartment: a multicisternal early endosome with multiple sorting defects. J Cell Sci. 2005;118:3003–17.
74 Baldys A, Raymond JR. Critical role of ESCRT machinery in EGFR recycling. Biochemistry. 2009;48:9321–3.
75 Guruharsha KG, Kankel MW & Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–66.
76 Moberg K, Schelble S, Burdick S, Hariharan I. Mutations in erupted, the Drosophila Ortholog of Mammalian Tumor Susceptibility Gene 101, Elicit Non-Cell-Autonomous Overgrowth. Dev Cell. 2005;9:699–710.
77 Thompson BJ, Mathieu J, Sung H-H, Loeser E, Rørth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–20.
78 Vaccari T, Bilder D. The Drosophila Tumor Suppressor vps25 Prevents Nonautonomous Overproliferation by Regulating Notch Trafficking. Dev Cell. 2005;9:687–98.
79 Herz H-M, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. Vps25 Mosaics Display Non-Autonomous Cell Survival and Overgrowth, and Autonomous Apoptosis. Development. 2006;133:1871–80.
80 Vaccari T, Rusten TE, Menut L, Nezis IP, Brech A, Stenmark H, Bilder D. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J Cell Sci. 2009;122:2413–23.
81 Tognon E, Wollscheid N, Cortese K, Tacchetti C, Vaccari T. ESCRT-0 is not required for ectopic Notch activation and tumor suppression in Drosophila. PLoS One. 2014;9:e93987.
82 Gilbert MM, Robinson BS, Moberg KH. Functional interactions between the erupted/tsg101 growth suppressor gene and the DaPKC and rbf1 genes in Drosophila imaginal disc tumors. PLoS One. 2009;4:e7039.
83 Palacios F, Tushir JS, Fujita Y, D’Souza-Schorey C. Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol Cell Biol. 2005;25:389–402.
84 Dukes JD, Fish L, Richardson JD, Blaikley E, Burns S, Caunt CJ, et al. Functional ESCRT machinery is required for constitutive recycling of claudin-1 and maintenance of polarity in vertebrate epithelial cells. Mol Biol Cell. 2011;22:3192–205.
85 Leithe E, Kjenseth A, Sirnes S, Stenmark H, Brech A, Rivedal E. Ubiquitylation of the gap junction protein connexin-43 signals its trafficking from early endosomes to lysosomes in a process mediated by Hrs and Tsg101. J Cell Sci. 2009;122:3883–93.
86 Lobert VH, Stenmark H. Cell polarity and migration: emerging role for the endosomal sorting machinery. Physiology (Bethesda). 2011;26:171–80.
87 Herz H-M, Woodfield SE, Chen Z, Bolduc C, Bergmann A. Common and distinct genetic properties of ESCRT-II components in Drosophila. PLoS One. 2009;4:e4165.
88 Woodfield SE, Graves HK, Hernandez JA, Bergmann A. De-regulation of JNK and JAK/STAT signaling in ESCRT-II mutant tissues cooperatively contributes to neoplastic tumorigenesis. PLoS One. 2013;8:e56021.
89 Vaccari T, Bilder D. At the crossroads of polarity, proliferation and apoptosis: the use of Drosophila to unravel the multifaceted role of endocytosis in tumor suppression. Mol Oncol. 2009;3:354–65.
90 Komada M, Soriano P. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 1999;13:1475–85.
91 Ruland J, Sirard C, Elia A, MacPherson D, Wakeham A, Li L, et al. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc Natl Acad Sci U S A. 2001;98:1859–64.
92 Yamada M, Ishii N, Asao H, Murata K, Kanazawa C, Sasaki H, Sugamura K. Signal-transducing adaptor molecules STAM1 and STAM2 are required for T-cell development and survival. Mol Cell Biol. 2002;22:8648–58.
93 Shim J-H, Xiao C, Hayden MS, Lee K-Y, Trombetta ES, Pypaert M, et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J Cell Biol. 2006;172:1045–56.
94 Handschuh K, Feenstra J, Koss M, Ferretti E, Risolino M, Zewdu R, et al. ESCRT-II/Vps25 constrains digit number by endosome-mediated selective modulation of FGF-SHH signaling. Cell Rep. 2014;9:674–87.
95 Rodahl LM, Haglund K, Sem-Jacobsen C, Wendler F, Vincent J-P, Lindmo K, et al. Disruption of Vps4 and JNK function in Drosophila causes tumour growth. PLoS One. 2009;4:e4354.
96 Huang H-R, Chen ZJ, Kunes S, Chang G-D, Maniatis T. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc Natl Acad Sci U S A. 2010;107:8322–7.
97 Seto ES, Bellen HJ. Internalization is required for proper Wingless signaling in Drosophila melanogaster. J. Cell Biol. 2006;173:95–106.
98 Lund VK, Delotto R. Regulation of Toll and Toll-like receptor signaling by the endocytic pathway. Small GTPases 2011;2:95–8.
99 Matusek T, Wendler F, Polès S, Pizette S, D’Angelo G, Fürthauer M, Thérond PP. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature. 2014;516:99–103.
100 Taelman VF, Dobrowolski R, Plouhinec J-L, Fuentealba LC, Vorwald PP, Gumper I, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–48.
101 Mamińska A, Bartosik A, Banach-Orłowska M, Pilecka I, Jastrzębski K, Zdżalik-Bielecka D, et al. ESCRT proteins restrict constitutive NF-κB signaling by trafficking cytokine receptors. Sci Signal. 2016;9:ra8-ra8.
102 Corless L, Crump CM, Griffin SDC, Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. 2010;91:362–72.
103 Jun M-H, Han J-H, Lee Y-K, Jang D-J, Kaang B-K, Lee J-A. TMEM106B, a frontotemporal lobar dementia (FTLD) modifier, associates with FTD-3-linked CHMP2B, a complex of ESCRT-III. Mol Brain. 2015;8:85.
104 Effantin G, Dordor A, Sandrin V, Martinelli N, Sundquist WI, Schoehn G, Weissenhorn W. ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cell Microbiol. 2013;15:213–26.
105 Bleck M, Itano MS, Johnson DS, Thomas VK, North AJ, Bieniasz PD, Simon SM. Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc Natl Acad Sci U S A. 2014;111:12211–6.
106 Van Engelenburg SB, Shtengel G, Sengupta P, Waki K, Jarnik M, Ablan SD, et al. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014;343:653–6.
107 Sandrin V, Sundquist WI. ESCRT requirements for EIAV budding. Retrovirology. 2013;10:104.
108 Prescher J, Baumgärtel V, Ivanchenko S, Torrano AA, Bräuchle C, Müller B, Lamb DCSuper-resolution imaging of ESCRT-proteins at HIV-1 assembly sites. PLoS Pathog. 2015;11:e1004677.
109 Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–42.
110 Pawliczek T, Crump CM. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol. 2009;83:11254–64.
111 Silvestri LS, Ruthel G, Kallstrom G, Warfield KL, Swenson DL, Nelle T, et al. Involvement of vacuolar protein sorting pathway in Ebola virus release independent of TSG101 interaction. J Infect Dis. 2007;196(Suppl):S264–70.
112 Madison MN, Okeoma CM. Exosomes: Implications in HIV-1 Pathogenesis. Viruses. 2015;7:4093–118.
113 Narayanan A, Iordanskiy S, Das R, Van Duyne R, Santos S, Jaworski E, et al. Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J Biol Chem. 2013;288:20014–33.
114 Kadiu I, Narayanasamy P, Dash PK, Zhang W, Gendelman HE. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. J Immunol. 2012;189:744–54.
115 Tamai K, Shiina M, Tanaka N, Nakano T, Yamamoto A, Kondo Y, et al. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology. 2012;422:377–85.
116 Temme S, Eis-Hübinger AM, McLellan AD, Koch N. The herpes simplex virus-1 encoded glycoprotein B diverts HLA-DR into the exosome pathway. J Immunol. 2010;184:236–43.
117 Alenquer M, Amorim M. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses. 2015;7:5066–83.
118 Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MAJ, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–33.
119 Meckes DG, Shair KHY, Marquitz AR, Kung C-P, Edwards RH, Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;10:20370–5.
120 Silva-Ayala D, López T, Gutiérrez M, Perrimon N, López S, Arias CF. Genome-wide RNAi screen reveals a role for the ESCRT complex in rotavirus cell entry. Proc Natl Acad Sci U S A. 2013;110:10270–5.
121 Li Z, Blissard GW. Cellular VPS4 is required for efficient entry and egress of budded virions of Autographa californica multiple nucleopolyhedrovirus. J Virol. 2012;86:459–72.
122 Shtanko O, Nikitina RA, Altuntas CZ, Chepurnov AA, Davey RA. Crimean-Congo hemorrhagic fever virus entry into host cells occurs through the multivesicular body and requires ESCRT regulators. PLoS Pathog. 2014;10:e1004390.
123 Pasqual G, Rojek JM, Masin M, Chatton J-Y, Kunz S. Old world arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS Pathog. 2011;7:e1002232.
124 Broniarczyk J, Bergant M, Goździcka-Józefiak A, Banks L. Human papillomavirus infection requires the TSG101 component of the ESCRT machinery. Virology. 2014;460–461, 83–90.
125 Garrison AR, Radoshitzky SR, Kota KP, Pegoraro G, Ruthel G, Kuhn JH, et al. Crimean-Congo hemorrhagic fever virus utilizes a clathrin- and early endosome-dependent entry pathway. Virology. 2013;444:45–54.
126 Simon M, Johansson C, Mirazimi A. Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH- and cholesterol-dependent. J Gen Virol. 2009;90:210–5.
127 Ghoujal B, Milev MP, Ajamian L, Abel K, Mouland AJ. ESCRT-II’s involvement in HIV-1 genomic RNA trafficking and assembly. Biol Cell. 2012;104:706–21.
128 Meng B, Ip NCY, Prestwood LJ, Abbink TEM, Lever AML. Evidence that the endosomal sorting complex required for transport-II (ESCRT-II) is required for efficient human immunodeficiency virus-1 (HIV-1) production. Retrovirology. 2015;12:72.
129 Stieler JT, Prange R. Involvement of ESCRT-II in hepatitis B virus morphogenesis. PLoS One. 2014;9:e91279.
130 Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–51.
131 Philips JA, Porto MC, Wang H, Rubin EJ, Perrimon N. ESCRT factors restrict mycobacterial growth. Proc Natl Acad Sci U S A. 2008;105:3070–5.
132 Mehra A, Zahra A, Thompson V, Sirisaengtaksin N, Wells A, Porto M, et al. Mycobacterium tuberculosis Type VII Secreted Effector EsxH Targets Host ESCRT to Impair Trafficking. PLoS Pathog. 2013;9:e1003734.
133 Abrami L, Brandi L, Moayeri M, Brown MJ, Krantz BA, Leppla SH, VanderGoot FG. Hijacking Multivesicular Bodies Enables Long-Term and Exosome-Mediated Long-Distance Action of Anthrax Toxin. Cell Rep. 2013;5:986–96.
134 Wolf JM, Johnson DJ, Chmielewski D, Davis DA. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot. Cell. 2010;9:1203–15.
135 Cornet M, Bidard F, Schwarz P, Da Costa G, Blanchin-Roland S, Dromer F, Gaillardin C. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect Immun. 2005;73:7977–87.
136 Kullas AL, Li M, Davis DA. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot Cell. 2004;3:1609–18.
137 Xu W, Smith FJ, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell. 2004;15:5528–37.
138 Zhang Y, Li W, Chu M, Chen H, Yu H, Fang C, et al. The AAA ATPase Vps4 Plays Important Roles in Candida albicans Hyphal Formation and is Inhibited by DBeQ. Mycopathologia. 2015;181:1–11.
139 Wolf JM, Davis DA. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics. 2010;184:673–94.
140 Li L, Cohen SN. tsg101: A Novel Tumor Susceptibility Gene Isolated by Controlled Homozygous Functional Knockout of Allelic Loci in Mammalian Cells. Cell. 1996;85:319–29.
141 Broniarczyk J, Olejnik-Schmidt AK, Luczak MW, Schmidt MT, Dabrowski M, Józefiak A, et al. Analysis of expression and structure of the TSG101 gene in cervical cancer cells. Int J Mol Med. 2010;25:777–83.
142 Krempler A, Henry MD, Triplett AA, Wagner K-U. Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53-independent cell death. J Biol Chem. 2002;277:43216–23.
143 Wagner K-U, Krempler A, Qi Y, Park K, Henry MD, Triplett AA, et al. Tsg101 is essential for cell growth, proliferation, and cell survival of embryonic and adult tissues. Mol Cell Biol. 2003;23:150–62.
144 Feng GH, Lih CJ, Cohen SN. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res. 2000;60:1736–41.
145 Oh KB, Stanton MJ, West WW, Todd GL, Wagner K-U. Tsg101 is upregulated in a subset of invasive human breast cancers and its targeted overexpression in transgenic mice reveals weak oncogenic properties for mammary cancer initiation. Oncogene. 2007;26:5950–9.
146 Liu F, Yu Y, Jin Y, Fu S. TSG101, identified by screening a cancer cDNA library and soft agar assay, promotes cell proliferation in human lung cancer. Mol Biol Rep. 2010;37:2829–38.
147 Liu D, Yang Z, Jiang S. Identification of PEG10 and TSG101 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Pathol Oncol Res. 2011;17:859–66.
148 Liu R-T, Huang C-C, You H-L, Chou F-F, Hu C-CA, Chao F-P, et al. Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas. Oncogene. 2002;21:4830–7.
149 Young TW, Mei FC, Rosen DG, Yang G, Li N, Liu J, Cheng X. Up-regulation of tumor susceptibility gene 101 protein in ovarian carcinomas revealed by proteomics analyses. Mol Cell Proteomics. 2007;6:294–304.
150 Zhui G, Gilchrist R, Borley N, Chng HW, Morgan M, Marshall JF, et al. Reduction of TSG101 protein has a negative impact on tumor cell growth. Int J Cancer. 2004;109:541–7.
151 Zhang Y, Song M, Cui ZS, Li CY, Xue XX, Yu M, Lu Y, et al. Down-regulation of TSG101 by small interfering RNA inhibits the proliferation of breast cancer cells through the MAPK/ERK signal pathway. Histol Histopathol. 2011;26:87–94.
152 Young TW, Rosen DG, Mei FC, Li N, Liu J, Wang X-F, Cheng X. Up-regulation of tumor susceptibility gene 101 conveys poor prognosis through suppression of p21 expression in ovarian cancer. Clin Cancer Res. 2007;13:3848–54.
153 Xu Z, Liang L, Wang H, Li T, Zhao M. HCRP1, a novel gene that is downregulated in hepatocellular carcinoma, encodes a growth-inhibitory protein. Biochem Biophys Res Commun. 2003;311:1057–66.
154 Lai MW, Huang SF, Lin SM, Chen TC, Lin CY, Yeh CN, et al. Expression of the HCRP1 mRNA in HCC as an independent predictor of disease-free survival after surgical resection. Hepatol Res. 2009;39:164–76.
155 Xu J, Yang W, Wang Q, Zhang Q, Li X, Lin X, et al. Decreased HCRP1 expression is associated with poor prognosis in breast cancer patients. Int J Clin Exp Pathol. 2014;7:7915–22.
156 Wittinger M, Vanhara P, El-Gazzar A, Savarese-Brenner B, Pils D, Anees M, et al. hVps37A Status affects prognosis and cetuximab sensitivity in ovarian cancer. Clin Cancer Res. 2011;17:7816–27.
157 Chen F, Deng J, Liu X, Li W, Zheng J. HCRP-1 regulates cell migration and invasion via EGFR-ERK mediated up-regulation of MMP-2 with prognostic significance in human renal cell carcinoma. Sci Rep. 2015;5:13470.
158 You Z, Xin Y, Liu Y, Sun J, Zhou G, Gao H, Xu P, et al. Chmp1A acts as a tumor suppressor gene that inhibits proliferation of renal cell carcinoma. Cancer Lett. 2012;319:190–6.
159 Li J, Belogortseva N, Porter D, Park M. Chmp1A functions as a novel tumor suppressor gene in human embryonic kidney and ductal pancreatic tumor cells. Cell Cycle. 2008;7:2886–93.
160 Mochida GH, Ganesh VS, de Michelena MI, Dias H, Atabay KD, Kathrein KL, et al. CHMP1A encodes an essential regulator of BMI1-INK4A in cerebellar development. Nat Genet. 2012;44:1260–4.
161 Hu B, Jiang D, Chen Y, Wei L, Zhang S, Zhao F, Ni R, et al. High CHMP4B expression is associated with accelerated cell proliferation and resistance to doxorubicin in hepatocellular carcinoma. Tumor Biol. 2015;36:2569–81.
162 Li K, Liu J, Tian M, Gao G, Qi X, Pan Y, et al. CHMP4C Disruption Sensitizes the Human Lung Cancer Cells to Irradiation. Int J Mol Sci. 2015;17.
163 Wei J, Lv L, Wan Y, Cao Y, Li G, Lin H, et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284–94.
164 Chen VY, Posada MM, Blazer LL, Zhao T, Rosania GR. The role of the VPS4A-exosome pathway in the intrinsic egress route of a DNA-binding anticancer drug. Pharm Res. 2006;23:1687–95.
165 Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, Howell SB. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4:1595–604.
166 Toyoshima M, Tanaka N, Aoki J, Tanaka Y, Murata K, Kyuuma M, et al. Inhibition of tumor growth and metastasis by depletion of vesicular sorting protein Hrs: Its regulatory role on E-cadherin and β-catenin. Cancer Res. 2007;67:5162–71.
167 Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–8.
168 van der Zee J, Urwin H, Engelborghs S, Bruyland M, Vandenberghe R, Dermaut B, et al. CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum Mol Genet. 2008;17:313–22.
169 Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Göttlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci U S A. 2006;103:19140–5.
170 Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–4.
171 Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–9.
172 Urwin H, Authier A, Nielsen JE, Metcalf D, Powell C, Froud K, et al. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19:2228–38.
173 Lee J-A, Beigneux A, Ahmad ST, Young SG, Gao F-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–7.
174 Nielsen TT, Mizielinska S, Hasholt L, Isaacs AM, Nielsen JE. Reversal of pathology in CHMP2B-mediated frontotemporal dementia patient cells using RNA interference. J Gene Med. 2012;14:521–9.
175 Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227.
176 Cox LE, Ferraiuolo L, Goodall EF, Heath PR, Higginbottom A, Mortiboys H, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS). PLoS One. 2010;5:e9872.
177 Holm IE, Englund E, Mackenzie IRA, Johannsen P, Isaacs AM. A reassessment of the neuropathology of frontotemporal dementia linked to chromosome 3. J Neuropathol Exp Neurol. 2007;66:884–91.
178 Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, Andersen PM, et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology. 2006;67:1074–7.
179 Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet. 2005;14:19–38.
180 Park SH, Zhu P-P, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120:1097–110.
181 Papadopoulos C, Orso G, Mancuso G, Herholz M, Gumeni S, Tadepalle N, et al. Spastin binds to lipid droplets and affects lipid metabolism. PLoS Genet. 2015;11:e1005149.
182 Zivony-Elboum Y, Westbroek W, Kfir N, Savitzki D, Shoval Y, Bloom A, et al. A founder mutation in Vps37A causes autosomal recessive complex hereditary spastic paraparesis. J Med Genet. 2012;49:462–72.
183 Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol. 1998;152:879–84.
184 Braak H, Tredici K Del, Rüb U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211.
185 Lee S-J, Lim H-S, Masliah E, Lee H-J. Protein aggregate spreading in neurodegenerative diseases: problems and perspectives. Neurosci Res. 2011;70:339–48.
186 Spencer B, Kim C, Gonzalez T, Bisquertt A, Patrick C, Rockenstein E, et al. α-Synuclein interferes with the ESCRT-III complex contributing to the pathogenesis of Lewy Body disease. Hum Mol Genet. 2016;25:1100–15.
187 Hasegawa T, Konno M, Baba T, Sugeno N, Kikuchi A, Kobayashi M, et al. The AAA-ATPase VPS4 regulates extracellular secretion and lysosomal targeting of α-synuclein. PLoS One. 2011;6:e29460.
188 Alvarez-Erviti L, Seow Y, Schapira AH, Gardiner C, Sargent IL, Wood MJA, Cooper JM. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol Dis. 2011;42:360–7.
189 Spencer B, Emadi S, Desplats P, Eleuteri S, Michael S, Kosberg K, et al. ESCRT-mediated uptake and degradation of brain-targeted α-synuclein single chain antibody attenuates neuronal degeneration in vivo. Mol Ther. 2014;22:1753–67.
190 Kurashige T, Takahashi T, Yamazaki Y, Hiji M, Izumi Y, Yamawaki T, Matsumoto M. Localization of CHMP2B-immunoreactivity in the brainstem of Lewy body disease. Neuropathology. 2013;33:237–45.
191 Edgar JR, Willén K, Gouras GK, Futter CE. ESCRTs regulate amyloid precursor protein sorting in multivesicular bodies and intracellular amyloid-β accumulation. J Cell Sci. 2015;128:2520–8.
192 Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EMC, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500.
193 Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–31.
194 Shiels A, Bennett TM, Knopf HLS, Yamada K, Yoshiura K, Niikawa N, Shim S, Hanson PI. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet. 2007;81:596–606.
195 Sagona AP, Nezis IP, Stenmark H. Association of CHMP4B and autophagy with micronuclei: implications for cataract formation. Biomed Res Int. 2014, 974393.
196 Zhang D, Wang L, Yan L, Miao X, Gong C, Xiao M, et al. Vacuolar protein sorting 4B regulates apoptosis of intestinal epithelial cells via p38 MAPK in Crohn’s disease. Exp Mol Pathol. 2015;98:55–64.
197 Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–407.
198 Komada M, Kitamura N. Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol Cell Biol. 1995;15:6213–21.
199 Lu L, Komada M, Kitamura N. Human Hrs, a tyrosine kinase substrate in growth factor-stimulated cells: cDNA cloning and mapping of the gene to chromosome 17. Gene. 1998;213:125–32.
200 Urbé S, Mills IG, Stenmark H, Kitamura N, Clague MJ. Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate. Mol Cell Biol. 2000;20:7685–92.
201 Stern KA, Visser Smit GD, Place TL, Winistorfer S, Piper RC, Lill NL. Epidermal growth factor receptor fate is controlled by Hrs tyrosine phosphorylation sites that regulate Hrs degradation. Mol Cell Biol. 2007;27:888–98.
202 Urbé S, Sachse M, Row PE, Preisinger C, Barr FA, Strous G, Klumperman J, Clague MJ. The UIM domain of Hrs couples receptor sorting to vesicle formation. J Cell Sci. 2003;116:4169–79.
203 Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8:163–9.
204 Row PE, Clague MJ, Urbé S. Growth factors induce differential phosphorylation profiles of the Hrs-STAM complex: a common node in signalling networks with signal-specific properties. Biochem J. 2005;389:629–36.
205 Kim BY, Olzmann JA, Barsh GS, Chin L-S, Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell. 2007;18:1129–42.
206 Jiao J, Sun K, Walker WP, Bagher P, Cota CD, Gunn TM. Abnormal regulation of TSG101 in mice with spongiform neurodegeneration. Biochim Biophys Acta. 2009;1792:1027–35.
207 Majumder P, Chakrabarti O. Mahogunin regulates fusion between amphisomes/MVBs and lysosomes via ubiquitination of TSG101. Cell Death Dis. 2015;6:e1970.
208 Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–52.
209 McDonald B, Martin-Serrano J. Regulation of Tsg101 expression by the steadiness box: a role of Tsg101-associated ligase. Mol Biol Cell. 2008;19:754–63.
210 Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, Miguet N, et al. CC2D1A is a regulator of ESCRT-III CHMP4B. J Mol Biol. 2012;419:75–88.
211 Usami Y, Popov S, Weiss ER, Vriesema-Magnuson C, Calistri A, Göttlinger HG. Regulation of CHMP4/ESCRT-III function in human immunodeficiency virus type 1 budding by CC2D1A. J Virol. 2012;86:3746–56.
212 >Drusenheimer N, Migdal B, Jäckel S, Tveriakhina L, Scheider K, Schulz K, et al. The Mammalian Orthologs of Drosophila Lgd, CC2D1A and CC2D1B, Function in the Endocytic Pathway, but Their Individual Loss of Function Does Not Affect Notch Signalling. PLoS Genet. 2015;11:e1005749.
213 Jaekel R, Klein T. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev Cell. 2006;11:655–69.
214 Troost T, Jaeckel S, Ohlenhard N, Klein T. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J Cell Sci. 2012;125:763–76.
215 Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev Cell. 2006;11:641–53.
216 Matias NR, Mathieu J, Huynh J-R. Abscission is regulated by the ESCRT-III protein shrub in Drosophila germline stem cells. PLoS Genet. 2015;11:e1004653.
217 Morawa KS, Schneider M, Klein T. Lgd regulates the activity of the BMP/Dpp signalling pathway during Drosophila oogenesis. Development. 2015;142:1325–35.
218 Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–9.
219 Palmer WH, Deng W-M. Ligand-Independent Mechanisms of Notch Activity. Trends Cell Biol. 2015;25:697–707.
220 Kobia F, Duchi S, Deflorian G, Vaccari T. Pharmacologic inhibition of vacuolar H+ ATPase reduces physiologic and oncogenic Notch signaling. Mol Oncol. 2014;8:207–20.
221 Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33.
222Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32.
223 Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108.
224 Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42.
225 Song W, Wang F, Lotfi P, Sardiello M, Segatori L. 2-Hydroxypropyl-β-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: Implications for therapy. J Biol Chem. 2014;289:10211–22.
226 Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Björklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110:E1817-26.
227 Bové J, Martínez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12,:437–52.
228 Kilpatrick K, Zeng Y, Hancock T, Segatori L. Genetic and chemical activation of TFEB mediates clearance of aggregated α-synuclein. PLoS One. 2015;10:e0120819.
229 Tognon E, Kobia F, Busi I, Fumagalli A, De Masi F, Vaccari T. Control of lysosomal biogenesis and Notch-dependent tissue patterning by components of the TFEB-V-ATPase axis in Drosophila melanogaster. Autophagy. 2016;12:499–514.
230 Miura GI, Roignant J-Y, Wassef M, Treisman JE. Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development. 2008;135:1913–22.
231 Parkinson MDJ, Piper SC, Bright NA, Evans JL, Boname JM, Bowers K, et al. A non-canonical ESCRT pathway, including histidine domain phosphotyrosine phosphatase (HD-PTP), is used for down-regulation of virally ubiquitinated MHC class I. Biochem J. 2015;471:79–88.
232 Ali N, Zhang L, Taylor S, Mironov A, Urbé S, Woodman P. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol. 2013;23:453–61.
233 Lee J, Oh K-J, Lee D, Kim BY, Choi JS, Ku B, Kim SJ. Structural Study of the HD-PTP Bro1 Domain in a Complex with the Core Region of STAM2, a Subunit of ESCRT-0. PLoS One. 2016;11:e0149113.
234 Pradhan-Sundd T, Verheyen EM. The Myopic-Ubpy-Hrs nexus enables endosomal recycling of Frizzled. Mol Biol Cell. 2015;26:3329–42.
235 Toyooka S, Ouchida M, Jitsumori Y, Tsukuda K, Sakai A, Nakamura A, et al. HD-PTP: A novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem Biophys Res Commun. 2000;278:671–8.
236 Gingras M-C, Zhang YL, Kharitidi D, Barr AJ, Knapp S, Tremblay ML, Pause A. HD-PTP is a catalytically inactive tyrosine phosphatase due to a conserved divergence in its phosphatase domain. PLoS One. 2009;4:e5105.
237 Haller O, Weber F. Pathogenic viruses: smart manipulators of the interferon system. Curr Top Microbio. Immunol. 2007;316:315–34.
238 Harty RN, Pitha PM, Okumura A. Antiviral activity of innate immune protein ISG15. J Innate Immun. 2009;1:397–404.
239 Morales DJ, Lenschow DJ. The antiviral activities of ISG15. J Mol Biol. 2013;425:4995–5008.
240 Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos N V, Lutz A, Wolff T, et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A. 2007;104:1371–6.
241 Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A. 2006;103:1440–5.
242 Pincetic A, Kuang Z, Seo EJ, Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010;84:4725–36.
243 Vild CJ, Li Y, Guo EZ, Liu Y, Xu Z. A novel mechanism of regulating the ATPase VPS4 by its cofactor LIP5 and the endosomal sorting complex required for transport (ESCRT)-III protein CHMP5. J Biol Chem. 2015;290:7291–303.
244 Shim S, Merrill SA, Hanson PI. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell. 2008;19:2661–72.
245 Skalicky JJ, Arii J, Wenzel DM, Stubblefield W-MB, Katsuyama A, Uter NT, et al. Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport. J Biol Chem. 2012;287:43910–26.
246 Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J Virol. 2003;77:6507–19.
247 Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002;99:955–60.
248 Chen H, Liu X, Li Z, Zhan P, De Clercq E. TSG101: a novel anti-HIV-1 drug target. Curr Med Chem. 2010;17:750–8.
249 Madara JJ, Han Z, Ruthel G, Freedman BD, Harty RN. The multifunctional Ebola virus VP40 matrix protein is a promising therapeutic target. Future Virol. 2015;10:537–546.
250 Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem Biol. 2008;3:757–64.
251 Martin-Serrano J, Neil SJD. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–31.
Disclosure statement: No financial support and no other potential conflict of interest relevant to this article was reported.