In vivo imaging of cardiac development and function in zebrafish using light sheet microscopy
DOI: https://doi.org/10.4414/smw.2015.14227
Michael
Weber, Jan
Huisken
Summary
Detailed studies of heart development and function are crucial for our understanding of cardiac failures and pave the way for better diagnostics and treatment. However, the constant motion and close incorporation into the cardiovascular system prevent in vivo studies of the living, unperturbed heart. The complementary strengths of the zebrafish model and light sheet microscopy provide a useful platform to fill this gap. High-resolution images of the embryonic vertebrate heart are now recorded from within the living animal: deep inside the unperturbed heart we can follow cardiac contractions and measure action potentials and calcium transients. Three-dimensional reconstructions of the entire beating heart with cellular resolution give new insights into its ever-changing morphology and facilitate studies into how individual cells form the complex cardiac network. In addition, cardiac dynamics and robustness are now examined with targeted optical manipulation. Overall, the combination of zebrafish and light sheet microscopy represents a promising addition for cardiac research and opens the door to a better understanding of heart function and development.
Key words: light sheet microscopy; zebrafish; in vivo imaging; embryonic heart; optical mapping; optogenetics
Introduction
The heart is an electrically controlled mechanical pump that persistently contracts and relaxes to move blood through the circulatory system. Malformation of cardiac structures during embryogenesis can quickly result in severe heart defects. Understanding development, morphology and function of the heart is therefore highly important and remains the focus of numerous studies. Due to the tight interrelation of structure and function of the heart and its close incorporation in the cardiovascular system, it should ideally be studied directly inside the body of the developing embryo. An understanding of the cardiac conduction system also requires the readout of electrical activity and calcium release, and strategies to manipulate it with high precision. Furthermore, deciphering the complex regulatory mechanisms of the cardiac muscle and revealing the cause of heart defects in early stages of development will require the ability to resolve individual heart cells and follow them with high temporal resolution over a long period of time.
However, such in vivostudies are often hindered by the physical characteristics of classic cardiovascular model organisms or limitations of the imaging technique. On the one hand, an embryo of the chosen model organism can be sacrificed at the developmental stage of interest and its heart excised, fixed, sectioned, stained and studied under a microscope [1, 2]. Although this method reveals valuable details about cardiac development, all the necessary experimental steps introduce artefacts that change the spatial relation of cardiac components and make measurements of size and distance prone to error [3]. On the other hand, current studies of cardiac dynamics typically require one to forgo high spatial resolution in order to achieve the necessary speed. Techniques such as magnetic resonance imaging, X-ray computed tomography or positron emission tomography provide exciting insights into living hearts, but cannot resolve details beyond tissue level [4, 5]. Moreover, functional studies are limited to electrophysiology of the excised heart or individual cardiac muscle cells. Therefore, various aspects of cardiac development, structure and function are still poorly understood.
Recently, the powerful combination of the model system zebrafish and light sheet microscopy has shown great promise for in vivostudies of heart development and function. The zebrafish has already been used as a model organism in cardiac research for more than two decades, but only with modern microscopy techniques are we finally able to exploit fully its benefits. With these developments, the embryonic zebrafish heart can now be recorded in vivo – in three dimensions (3D), with cellular resolution and, if required, even over the course of several hours or days. This opens the door for real-time developmental biology, where cardiac structure and function are studied unperturbed and in situ. Additionally, with the help of optical manipulation and optogenetic tools, researchers are able to not only monitor the heart, but also actively influence its function.
The zebrafish is a powerful model system for cardiac research
The zebrafish (Danio rerio) is native to rivers of Southeast Asia, reaches 4 cm in length and lives up to 5 years (fig. 1a). It has a high reproduction rate and can yield a high number of progeny: Mature female zebrafish lay up to 300 eggs per week, which are externally fertilised by males. As a result of its external development and optical clarity during embryogenesis, the zebrafish embryo is accessible for light microscopy studies from its earliest stages. Basic parameters such as heart rate, contractility and blood flow can be obtained by means of video microscopy [6]. Studying development and function of cardiovascular structures in vivo is facilitated by the ability to generate transgenic zebrafish expressing fluorescent proteins under the control of specific promoters [7–9].
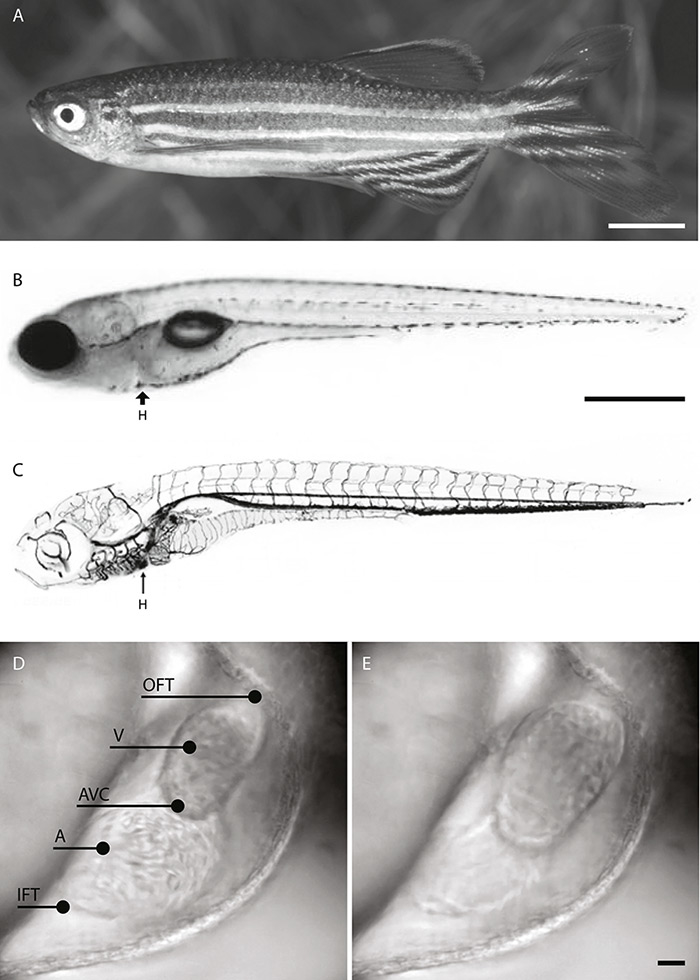
Figure 1
A: Adult male zebrafish. Scale bar 5 mm. B: Bright-field image of a 7 dpf (days post-fertilisation) zebrafish larva. Scale bar: 500 µm. C: The position of the heart (H) in the cardiovascular system of a zebrafish larva. Images adapted from Brown University (Isogai et al. 2001 [66]) and Interactive Atlas of Zebrafish Vascular Anatomy (zfish.nichd.nih.gov). D–E: Bright-field images of the 2 dpf zebrafish heart. D: Systole. E: Diastole. Inflow tract (IFT), atrium (A), atrioventricular canal (AVC), ventricle (V), outflow tract (OFT). Scale bar: 20 µm.
The cardiac development of the zebrafish shows remarkable similarities to that of humans. Less than 2 days after fertilisation, the zebrafish heart has developed from progenitor cells into a two-chambered organ that sits dorsally between the head and trunk (fig. 1b–c). The two chambers, atrium and ventricle, are connected by the atrioventricular canal (fig. 1d–e) and consist of two main cell layers: the inner endocardium and the outer myocardium. A double-walled sac, the pericardium, contains the heart and the roots of the great vessels while also fixing the heart to the thorax, providing lubrication and protecting against infection. In contrast to those of mammals and amphibians, the zebrafish heart does not progress to septation and retains a simpler structure. However, there are broad similarities between zebrafish and mammals with respect to the genetic determinants of heart tube and chamber formation [10].
Despite the comparatively simple structure of the zebrafish heart, its electrocardiogram and the overall shape of its action potentials are very similar to those of mammalian hearts [11–13]. A heart rate close to that of humans is maintained by its cardiac conduction system, which rapidly develops from a slow-conducting myocardial grid into a complex system with atrioventricular delay and fast ventricular apex-to-base activation [14].
The zebrafish is a powerful model system for in vivo studies of the genetic, morphological and functional basis of cardiovascular development. Large-scale genetic screens in zebrafish have identified numerous heart-specific mutations that are responsible for abnormal cardiovascular developments such as valve defects and aortic coarctation [10, 15–17]. Various mechanisms involved in human diseases can be modelled and studied in zebrafish, such as cardiomyopathy [18–20] and thrombosis [21, 22]. The known response to cardiac drugs such as astemizole and terfenadine can likewise be successfully reproduced in zebrafish [13, 23].
In contrast to other model organisms, zebrafish do not require active blood circulation during the first week after fertilisation. Embryos with cardiovascular defects that have a negative impact on blood flow can therefore continue development and facilitate longer phenotypic studies compared with mammals [24–26]. Moreover, the adult zebrafish heart has the ability to regenerate after the loss of up to 20% of its myocardium [27, 28]. Studying this astounding process in the living zebrafish heart may help to explain the inability of higher organisms to regenerate the heart [7, 29].
Overall, the zebrafish has unique features that make it attractive and complementary to existing model systems. The early presence of a beating heart and an advanced vasculature enables much more complex studies than cell or organ cultures. As a caveat, the smaller scale of the zebrafish heart and vasculature can result in relations that are different from those of higher model systems. For example, in comparison with the volume of the cardiac chambers and the diameter of blood vessels, erythrocytes are relatively large. Therefore, in some cases, inference to similar processes in higher organisms should be made with caution [30].
Light sheet microscopy is ideal for in vivo cardiac imaging
Despite the zebrafish embryo being well accessible for microscopy, imaging its heart with high resolution is very demanding. The primary reasons for this are its substantial size of about 250 µm and a heart rate of 2–4 Hz. Structural and functional cardiac imaging in the living zebrafish require a microscopy technique that records optical sections with high spatial and temporal resolution. In contrast to conventional light microscopy techniques, light sheet microscopy fulfils these requirements.

Figure 2
A: The principle of light sheet microscopy. Top: Illumination and detection objectives are oriented orthogonally, and the sample is placed at the intersection of their focal planes. A sheet of laser light illuminates a thin slice of the sample. Bottom: View from top. The light sheet has a waist in the centre of the field of view and overlaps perfectly with the focal plane of the detection objective. B: Custom-built light sheet microscope for imaging and photomanipulation of the zebrafish heart. C: Close-up of the medium-filled sample chamber with a blue light sheet. D: Sample mounting for in vivo cardiac optical mapping. Comparison of zebrafish embryos mounted in fluorinated ethylene propylene (FEP) tube and glass capillary. E: 1 dpf (days post-fertilisation) zebrafish mounted in FEP tube. F: Rendering of a mounted zebrafish embryo held by the sample holder inside the imaging chamber with illumination objectives, light sheet and detection objective.
Light sheet microscopy was introduced to the life sciences as a versatile fluorescence microscopy technique called selective plane illumination microscopy (SPIM) in 2004 [31]. The basic principle is intriguingly simple: a thin sheet of laser light illuminates a fluorescently labelled specimen from the side and excites fluorescence only in the focal plane of the detection objective while the emission light is recorded with a fast camera (fig. 2a) [32]. Light sheet microscopy has proved to be a powerful technique especially for biologists interested in imaging developmental processes in 3D. The key feature of light sheet microscopy – fast and gentle optical sectioning even with low magnification and low numerical aperture objectives – is a major improvement over classical fluorescence microscopy techniques. Another benefit of light sheet microscopy is the dramatically reduced energy input, as all light input contributes to useful signal. Now, even larger specimens such as entire organisms are imaged in real time and without the severe photo-toxicity and -bleaching effects known from previous fluorescence microscopy techniques [33, 34].
Since the first implementation of light sheet microscopy for developmental biology, numerous instrumental designs have been developed. Most of them share the same fundamental differences from traditional light microscopes. Light sheet microscopes are typically computer-controlled setups and come without traditional eyepieces, objective turret or xy-stage (fig. 2b). Often, the specimen sits in a dedicated vertical mount and is immersed in a medium-filled imaging chamber (fig. 2c). This facilitates rotation of the fragile sample without deformation and provides unobstructed views from all angles [33]. For imaging of zebrafish embryos, the temperature of the surrounding medium can also be adjusted to the zebrafish's preferred temperature of 28.5 °C.
Adjusted sample preparation is a crucial part to light sheet microscopy
In order to make full use of the new capabilities of light sheet microscopy, the sample preparation needs to be radically different from the dish or slide mounting used for conventional microscopy. Rather than being placed on a flat, horizontally oriented surface, the specimen is now held in a 3D environment that provides access from different angles and mimics its natural environment as closely as possible. For in vivolight sheet microscopy of zebrafish, the specimen has commonly been embedded in a low-melting-point agarose cylinder [31, 35]. The transparent agarose matches the refractive index of water and biological tissue, and concentrations of about 1.5% provide sufficient mechanical stability. To mount the sample, it is first immersed in melted agarose, then taken up into a glass capillary, and finally extruded from the capillary (fig. 2d). A zebrafish embryo mounted in solid agarose can be reproducibly translated and rotated to illuminate and image the heart or – another feature of interest – from the angle giving the optimum optical accessibility [3].
With use of light sheet microscopy, zebrafish development can now be imaged over longer periods of many hours or even several days [36, 37]. In contrast to specimens such as fruit fly embryos, the zebrafish continuously grows in size and substantially changes in shape during embryogenesis. Therefore, extended imaging requires further optimisation of the mounting technique, providing enough clearance for the sample to grow while keeping it in a semi-fixed position for precise microscopy recordings. Low-concentration agarose or methylcellulose, surrounded by a transparent polymer tube as mechanical support, has become a widely accepted solution. A well-known polymer is fluorinated ethylene propylene (FEP). It has near identical optical properties with water, and images are recorded right through it, while the plastic tube provides mechanical support to translate and rotate the sample (fig. 2d–f). In addition, the viscous liquid medium allows the fish to grow and develop normally inside the tube. In this manner, embryonic development of the zebrafish cardiovascular system can now be studied over the course of several days [36].
Optical sections of the zebrafish heart are recorded in real time
The instantaneous optical sectioning capabilities of light sheet microscopy are particularly valuable for in vivo cardiac imaging. Using the flexible sample orientation, the heart is precisely oriented in the field of view. Specific sections of the heart are isolated from surrounding tissue, and high contrast images are recorded inside the living zebrafish. Using a fast camera, the onset and progression of natural cardiac contractions from inflow to outflow tract are visualised and measured. Individual cells are identified by use of the high spatial resolution, and several layers of the heart like myocardium and endocardium are distinguished using tissue-specific fluorescence reporters (fig. 3a).
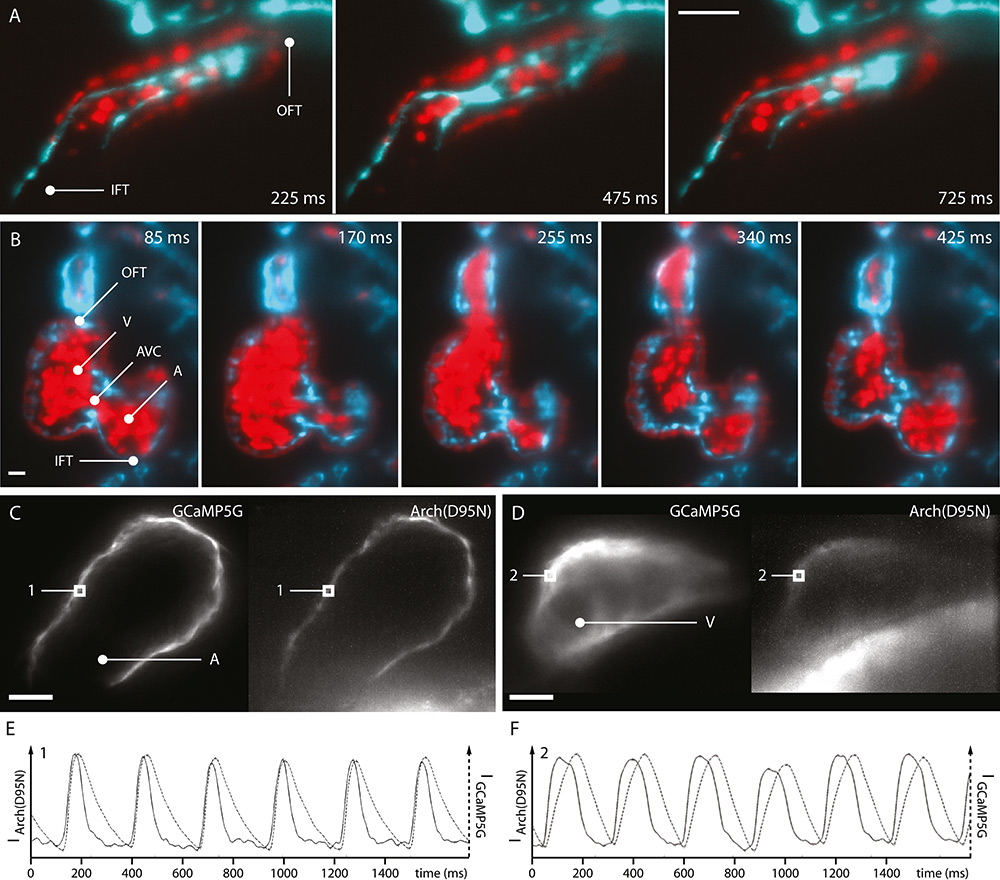
Figure 3
Optical sections of embryonic zebrafish hearts recorded with light sheet microscopy. A: 1 dpf transgenic Tg(kdrl:eGFP,myl7:dsRed, gata1a:dsRed) zebrafish. B: 4 dpf. C: Voltage and calcium imaging in the atrial myocardium of 2 dpf transgenic Tg(myl7:GCaMP5G-Arch(D95N) zebrafish, tnnt2a morpholino-injected. D: Ventricular myocardium. E: Action potentials and calcium transients in the atrial myocardium (position 1). F: Ventricular myocardium (position 2). Inflow tract (IFT), atrium (A), atrioventricular canal (AVC), ventricle (V), outflow tract (OFT). Scale bar: 20 µm.
In later stages of cardiac development, the formation and dynamic movement of the atrioventricular valve during heart contractions are visualised. Images depict the efficient closing of the valve during ventricular systole, and the quick opening during diastole. Blood flow is analysed by use of additional fluorescent labelling of blood cells. Even at the high acquisition speeds, optical sections of the zebrafish heart at this developmental stage reveal further details, such as ventricular trabeculation (fig. 3b).
Light sheet microscopy is also capable of cardiac functional imaging beyond the visualisation of the movements and morphology of the heart. Here, the combination of high speed imaging of optical sections with myocardial expressed voltage- and calcium-sensitive fluorescent proteins reveals the close correlation between action potentials and calcium transients. Those signals are simultaneously measured in vivoin all cells of the myocardial network visible in the images (fig. 3c–f). Their analysis provides an insight into the dynamics of cardiac control and reveals that action potentials and calcium transients in the zebrafish heart are chamber-specific [14].
The entire heart is imaged with cellular resolution
Selected regions of the heart can now be captured in vivo with great detail using light sheet microscopy. Although high-speed movies of a single plane are sufficient to describe and quantify numerous cardiac properties, they lack the depth information needed to reconstruct the entire beating heart. However, its continuous motion in all three dimensions makes it challenging to record high-resolution images of the entire organ. In a conventional light sheet microscope, moving the sample through the focal plane of the detection objective and consecutively recording adjacent planes of a z-stack is required to obtain 3D image data. With a fast microscope setup, capturing such a z-stack takes less than a second. But even when recorded at such high speed, rapid cardiac movements will distort the 3D image data and make precise measurements impossible. Nonetheless, several methods have been developed to solve this problem.
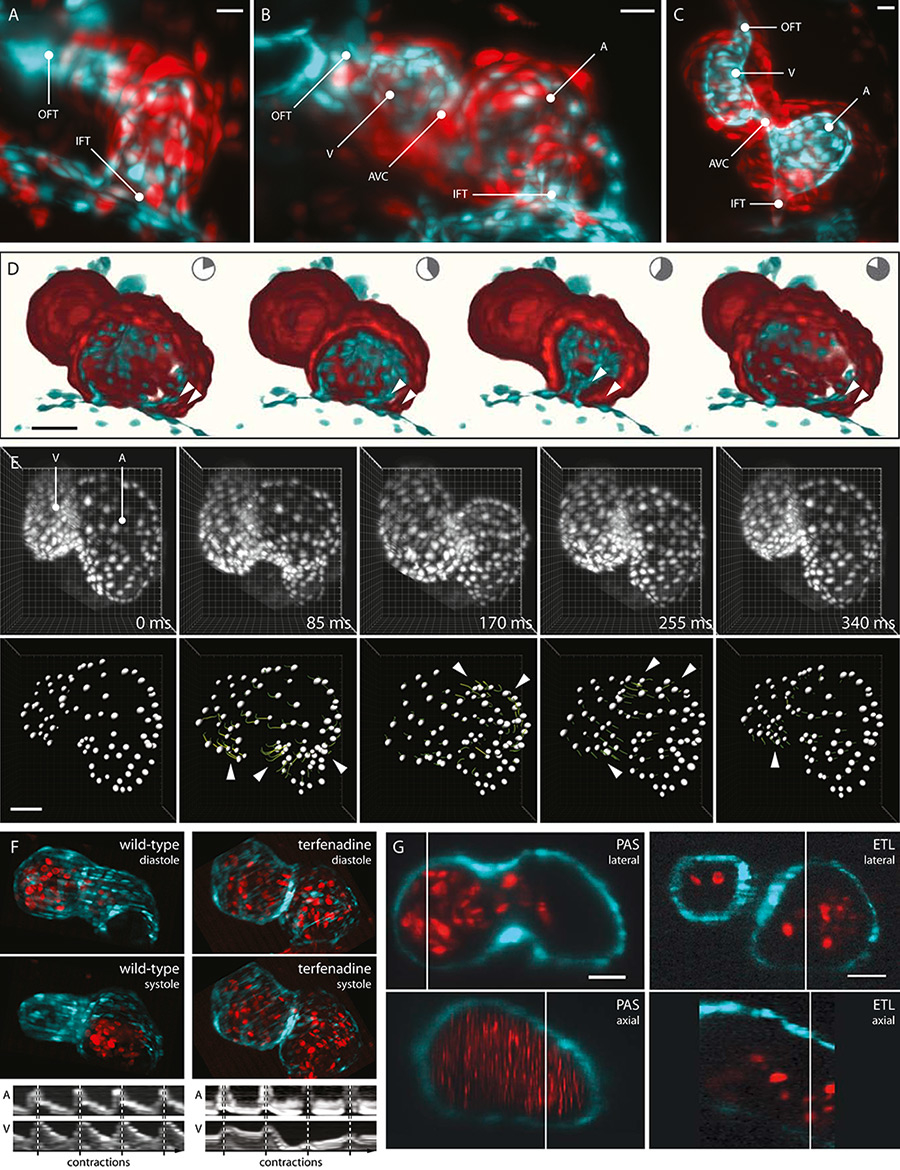
Figure 4
3D recordings of the embryonic zebrafish heart. A–C: Maximum intensity projections of image stacks recorded from silent hearts of 1–3 dpf transgenic Tg(kdrl:eGFP,myl7:dsRed, gata1a:dsRed) zebrafish. Inflow tract (IFT), atrium (A), atrioventricular canal (AVC), ventricle (V), outflow tract (OFT). Scale bar: 20 µm. D: 3D reconstruction of the beating heart of a 2 dpf zebrafish embryo. Cut open to reveal inflow tract and impact of cardiac jelly on pumping efficiency (arrow heads). E: 3D cell tracking in a beating zebrafish heart. 3D-projections of 2 dpf Tg(myl7:GCaMP5G-Arch(D95N), myl7:H2A-mCherry) zebrafish hearts. Postacquisition synchronisation, tracking based on segmentation of H2A-mCherry channel. Segmented cardiomyocytes with partial tracks (dragon tails, colour-coded for velocity). Areas of fastest contraction highlighted (arrow heads). F: Wild-type and arrhythmic hearts of 2 dpf transgenic Tg(myl7:GFP, GATA1:dsRed) zebrafish. Maximum intensity projections of rapid volume scanning image data. Arrhythmia induced by treatment with terfenadine. Kymographs of atrial and ventricular contractions. Ventricular contractions are absent in several cycles of the terfenadine-treated hearts. G: Comparison of 3D image data generated with postacquisition synchronisation (PAS) and rapid volume scanning (ETL). Hearts of 2 dpf of transgenic Tg(myl7:GFP, GATA1:dsRed) zebrafish. 3D blood cells are only visible in ETL data. Scale bar: 20 µm.
The first solution is to suppress the heartbeat: A mutation or blocked transcription of cardiac troponin T type 2a (tnnt2a) results in a “silent heart”. Combined with the unique ability of the zebrafish embryo to survive without circulation, z-stacks from silent hearts are then imaged throughout embryogenesis without any special technique (fig. 4a–c). Nevertheless, while facilitating a number of studies during embryogenesis, the silent heart phenotype has a visible impact on heart structure and development. Forces applied on the heart by cardiac contraction itself are crucial for proper formation of atrium and ventricle [38], and a working blood circulation is required for shaping the heart chambers [39, 40]. Therefore, “silent hearts” have only limited use in representative studies of cardiac development.
Alternatively, a static 3D image of the heart can be obtained with prospective gating methods. While the beating heart is moved step by step through the focal plane, it is continuously monitored using bright-field microscopy. In every plane, the recording of a fluorescent image is triggered at a predefined phase in the cardiac cycle. The recorded image data is afterwards reconstructed into a 3D model of the heart at this phase [41]. This method is useful if there is a certain cardiac phase of interest, but the dynamics of the heartbeat cannot be studied.
One technique with which three-dimensional cardiac dynamics are studied is postacquisition synchronisation with high-speed recording. Here, the light sheet microscope captures a z-stack of movies, with each movie covering at least one cardiac cycle. After the recording is finished, one 3D cardiac cycle is reconstructed by synchronising the movies in time. The result is image data that offer pristine views of the beating zebrafish heart [3].
Postacquisition synchronisation results in image data that offers new insights into the efficiency of the heart as a mechanical pump. High-speed imaging of a 3-day-old zebrafish atrium reveals the interplay between the contracting myocardium and the noncontracting cardiac jelly and endocardium. Comparatively slight myocardial contractions are amplified by the displacement of cardiac jelly and endocardium, resulting in almost complete atrial emptying (fig. 4d). Furthermore, cardiac contractions are now analysed in great detail using the cellular resolution of the resulting four-dimensional (4D, 3D + time) image data. The complex movement of the myocardium during systole and diastole and the propagation of cardiac contraction from inflow to outflow tract are visualised by tracking individual heart cells (fig. 4e).
Despite all the possibilities, all previously mentioned techniques share one limitation: 3D image data from nonperiodic movements, such as arrhythmic hearts or haemodynamics, cannot be recorded. It became necessary to further increase the acquisition speed in light sheet microscopy: In rapid volume scanning, instead of moving the specimen through the focal plane, the focal plane is moved through the specimen. The incorporation of an electrical tuneable lens as a remote focusing system and a synchronised scan mirror eliminate the need to move the sample or any heavy microscope component, thus enabling scan speeds that are beyond the reach of conventional volumetric imaging methods [42]. The entire heart is now instantaneously captured in rapid succession, and nonperiodic phenomena are studied with video-rate 3D imaging [3].
Rapid volume scanning delivers valuable images of fast irregular movements. As an example, arrhythmic hearts in embryos treated with terfenadine are imaged in 3D. The recorded image data reveal partially dropped ventricular systole and could be used to measure chamber volume or cell movement over time (fig. 4f). In contrast to the other techniques, rapid volume scanning is also capable of visualising individual blood cells in 3D as they are pumped through atrium and ventricle (fig. 4g).
Cardiac excitability is manipulated with light
The combination of zebrafish and light sheet microscopy provides unique insights into the unperturbed vertebrate heart. An enticing next step is to go beyond mere observational microscopy to influence actively cardiac activity. Optical manipulation offers precise perturbation of individual regions or the entire heart. Rather than manipulating the heart mechanically and risking severe side effects, optical manipulation tends to be more precise and gentle. For example, selective photoconversion of fluorescent proteins is used to trace the lineage of individual cells [43]. To compare the robustness and behaviour of the heart in healthy and diseased states, optogenetic tools are used to manipulate precisely and reversibly cardiac excitability [44]. These experiments demonstrate that light-activated hyper- and hypopolarisation can replace conventional electrical stimulation and even overcome some of the known limitations such as short activation times and alterations of pH [45].
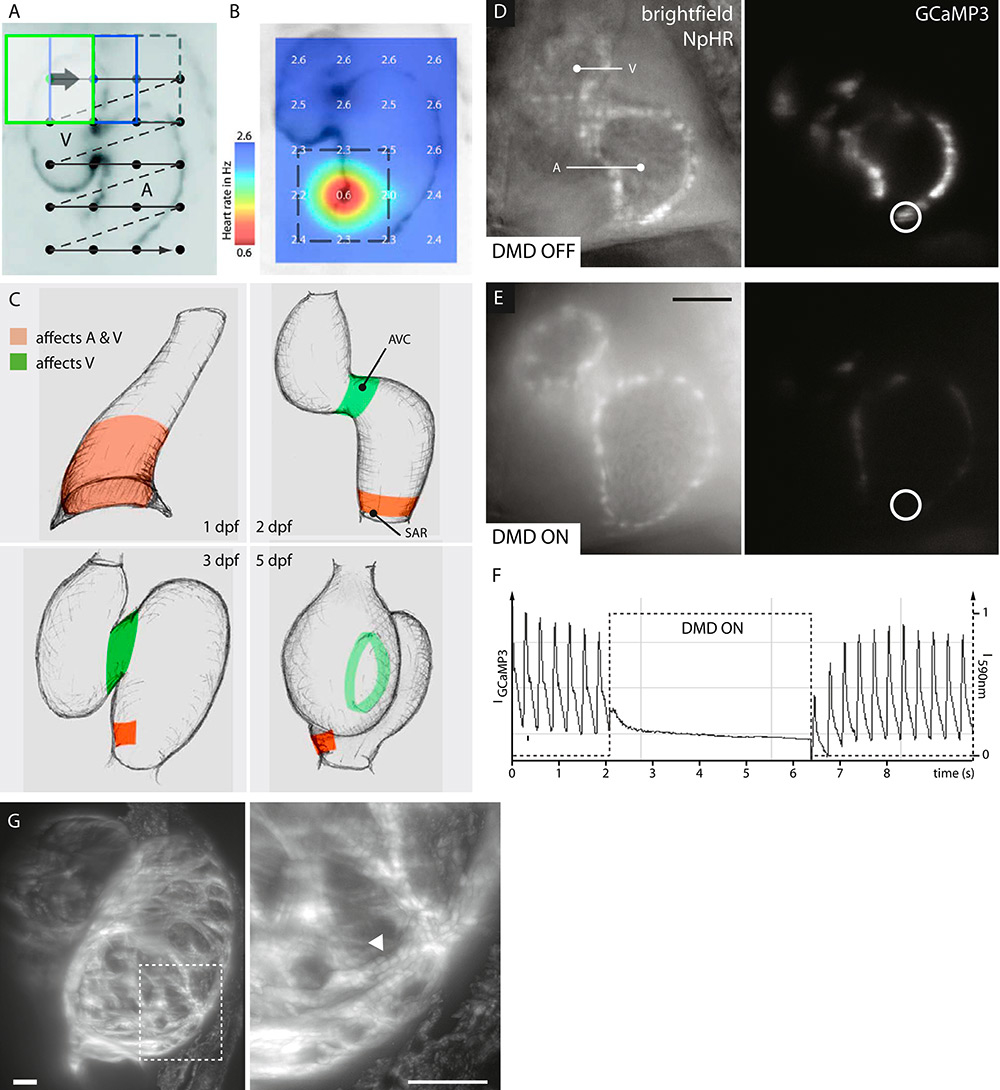
Figure 5
Optical manipulation in halorhodopsin-expressing zebrafish hearts. A-C: Patterned illumination reveals the location of the sinus venosus in hearts of transgenic Tg(s1101t:Gal4, UAS:NpHR-mCherry, UAS:Kaede) zebrafish. A: A sequence of rectangular 590 nm illumination is moved over the zebrafish heart located in the field of view. B: A heat map of the heart rate is generated from simultaneously recorded movies, highlighting the area with the highest impact on overall heart rate. C: Areas that control atrial and ventricular (red) or only ventricular (green) contractions are revealed throughout embryogenesis. D–F: Optical manipulation and simultaneous calcium imaging reveal myocardial calcium levels. 3 dpf transgenic Tg(myl7:Gal4, UAS:NpHR-mCherry, UAS:GCaMP3) zebrafish. D: Unperturbed heart with periodic cardiac contractions and calcium release. E: Optogenetically silenced heart without calcium release. F: Calcium transients before, during and after optical manipulation. A saturation effect is visible upon release of illumination. Scale bar: 50 µm. G: High-resolution 3D imaging of sarcomere structures (arrow head) in an optically relaxed heart. Transgenic Tg(myl7:LifeAct-GFP, myl7:Gal4, UAS:NpHR-mCherry) zebrafish. 5 s illumination with 590 nm and simultaneous image recording.
Light sheet microscopy is a practical basis for optical manipulation in the zebrafish heart. Custom-built setups provide the necessary flexibility to integrate optical manipulation without hindering the imaging capabilities of the microscope [46]. Recently, the combination of light sheet microscopy and optical manipulation was used to locate the sinus venosus of the zebrafish. The heart rate was monitored while illuminating different regions of zebrafish hearts expressing the light-controlled chloride pump halorhodopsin [47] in their myocardial membrane. The activation of halorhodopsin hyperpolarises the myocardium and suppresses the formation of action potentials, resulting in a reduced or absent heart rate (fig. 5a-b). The sinus venosus could subsequently be identified by a cardiac arrest when illuminated. Over the course of embryogenesis, the area of primary pacemaker activity decreases and a region of ventricular pacemaker activity could be located at the atrioventricular canal at 2 days after fertilization (fig. 5c) [48].
Optical manipulation becomes especially powerful when combined with fluorescent reporters of cardiac activity. Cardiac optical mapping visualises action potentials and calcium transients by utilising voltage- and calcium-sensitive fluorescent dyes or genetically encoded fluorescent proteins. This has proven to be a powerful tool for studying cardiovascular function and disease, cardiac conduction and electromechanical coupling [49–51]. New fluorescent reporters for optical mapping are developed at a rapid pace. One popular example is the genetically encoded calcium indicator GCaMP, which is created from a fusion of green fluorescent protein (GFP), calmodulin, and M13, a peptide sequence from myosin light chain kinase. Calcium binding to the calmodulin results in a structural shift and an increase in fluorescence intensity [52]. In zebrafish cardiac research, it has been used for studying arrhythmias [12], the development of the cardiac conduction system [14], the impact of cardiac conduction on heart chamber morphology [38] and the role of a Popeye domain containing genes for heart muscle development [53]. Genetically encoded voltage reporters such as Mermaid [54] and Arch(D95N) [55] have just recently been deployed for cardiac research [56, 57]. In contrast to the established optical mapping of cardiac action potentials using fluorescent dyes such as di-8-ANEPPS and Fura-2AM on excised hearts [58], genetically encoded fluorescent reporters are suited for in-vivo studies: They do not suffer from limited penetration, they do not display cytotoxic effects or get washed out, and they are tissue specific [59].
The onset of calcium release in the myocardium can now be monitored in transgenic zebrafish that simultaneously express a fluorescent calcium reporter such as GCaMP3 [60] and the light-controlled chloride pump halorhodopsin. In an unperturbed heart, periodic myocardial calcium release causes cardiac contractions (fig. 5c). As soon as the heart is illuminated with orange light, halorhodopsin hyperpolarises the myocardial membrane, the calcium concentration drops and cardiac contraction is diminished. Upon removal of illumination, the myocardial calcium concentration increases until it reaches saturation and cardiac contraction resumes immediately (fig. 5d–f).
Optical suppression of cardiac excitability is also a unique method to study quickly the otherwise consistently beating vertebrate heart in a fully relaxed state. The illumination of a halorhodopsin-expressing zebrafish heart results in immediate relaxation, and a 3D stack is recorded in the absence of any cardiac movement. Again, when the illumination ceases, the heart continues beating. This elegant solution does not impair cardiac development, while still providing high-resolution 3D image data (fig. 5g). Although the resulting images represent a normally developed heart, the depicted simultaneous diastole of atrium and ventricle obviously does not appear in normal cardiac cycles.
Overall, cardiac excitability is successfully controlled in vivo by optical manipulation, thanks to the optical accessibility of the zebrafish heart. In the future, this technique can also be useful to control hemodynamic forces during studies of heart formation or blood vessel development.
Conclusion
Studying the embryonic zebrafish with light sheet microscopy reveals previously unseen details of the vertebrate heart. This combination of a young cardiac model organism and a novel microscopy technique complements the established model systems and imaging technologies. Now, the heart can be studied in vivo at high speed with cellular resolution. The fast optical sectioning capabilities of light sheet microscopy reveal high-contrast images of cardiac contractions, action potentials and calcium signals in real time. The entire beating heart is now reconstructed using dedicated post-processing or near instantaneous volume scanning techniques. The recorded image data will be useful to analyse even subtle deviations between healthy and diseased hearts. Complemented by additional techniques such as optical manipulation, light sheet microscopy will help to understand the robustness of the heart and the contribution of individual cells to the complex cardiac network. Within the possibilities defined by the intrinsic limitations of zebrafish as a cardiac model system, many new insights into the heart and cardiovascular system will be gained.
A couple of technical challenges remain. Like every other light microscopy technique, light sheet microscopy suffers from light attenuation through absorption, scattering and refraction. This results in a decrease of contrast and spatial resolution in deeper regions of the embryonic zebrafish heart. Recently, the use of two-photon light sheet microscopy to improve penetration depth has been demonstrated in the zebrafish heart [61]. However, two-photon excitation still suffers from higher photobleaching, lower efficiency and lower frame rates when compared with single-photon light sheet microscopy [62]. Adaptive optics is an intriguing approach to correct for tissue-induced light attenuation; unfortunately, it remains far too slow for imaging fast dynamic processes such as the beating heart [63]. Another major challenge is that because light sheet microscopy produces high-resolution images at high speed, it also generates large amounts of image data. These data need to be transferred, stored and analysed, which requires much more attention to computational solutions than previous microscopy techniques. Intelligent data handling and real-time image analysis have been demonstrated for other tasks and could also be adapted to future cardiac imaging [64].
Long-term imaging of zebrafish development is now possible, thanks to dedicated sample mounting that allows the embryo to grow while keeping it in place for microscopy. Ideally, the zebrafish should be freely swimming and only stopped and kept in place for a quick image recording. Automated setups using camera-controlled robotic arms or pumps might even allow images from several fish to be recorded in parallel. The integration of optical manipulation beautifully demonstrates how microscopy goes beyond the mere recording of images. In the future, additional modalities such as electrophysiology or ultrasound imaging might further extend the capabilities of powerful microscopes. Ultimately, studies that are now carried out in embryos should also be possible in older zebrafish. With the development of pigment-free zebrafish mutants, the optically clear adult zebrafish has become a reality [65] and we may soon be able to record well-resolved images from within the adult zebrafish heart. Overall, light sheet microscopy has become the tool of choice for in vivo imaging and manipulation of the zebrafish heart. The combination thereof has the potential to bridge the gap between single cell studies and research on larger and more complex hearts.
References
1 Ieda M, et al. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat Med. 2007;13:604–12.
2 Ahmad F, et al. The role of cardiac troponin T quantity and function in cardiac development and dilated cardiomyopathy. PLoS ONE. 2008;3:e2642.
3 Mickoleit M, et al. High-resolution reconstruction of the beating zebrafish heart. Nat Methods. 2014;11:919–22.
4 Dzyubachyk, O, et al. Super-resolution reconstruction of late gadolinium-enhanced MRI for improved myocardial scar assessment. J Magn Reson Imaging. 2015;42:160–7.
5 Lapa C, et al. Imaging of myocardial inflammation with somatostatin receptor based PET/CT – A comparison to cardiac MRI. Int J Cardiol. 2015;194:44–9.
6 Schwerte T, Pelster B. Digital motion analysis as a tool for analysing the shape and performance of the circulatory system in transparent animals. J Exp Biol. 2000;203:1659–69.
7 Zhang R, et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature. 2013;1–6 doi:10.1038/nature12322
8 Rohr S, Otten C, Abdelilah-Seyfried S. Asymmetric Involution of the Myocardial Field Drives Heart Tube Formation in Zebrafish. Circ Res. 2008;102:e12–e19
9 Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev Biol. 2002;248:307–18.
10 Stainier DY. R. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48.
11 Nemtsas P, Wettwer E, Christ T, Weidinger G, Ravens U. Adult zebrafish heart as a model for human heart? An electrophysiological study. J Mol Cell Cardiol. 2010;48:161–71.
12 Arnaout R, et al. Zebrafish model for human long QT syndrome. Proc Natl Acad Sci USA. 2007;104:11316–21.
13 Milan DJ, Jones IL, Ellinor PT, Macrae CA. In vivo recording of adult zebrafish electrocardiogram and assessment of drug-induced QT prolongation. Am J Physiol Heart Circ Physiol. 2006;291:H269–73
14 Chi NC, et al. Genetic and Physiologic Dissection of the Vertebrate Cardiac Conduction System. PLoS Biol. 2008;6:e109.
15 Chen JN, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302.
16 Stainier DY, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–92.
17 Kikuchi Y, et al. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14:1279–89.
18 Xu X, et al. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–9.
19 Schönberger J, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37:418–22.
20 Gerull B, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30:201–4.
21 Gregory M, Hanumanthaiah R, Jagadeeswaran P. Genetic analysis of hemostasis and thrombosis using vascular occlusion. Blood Cells Mol Dis. 2002;29:286–95.
22 Jagadeeswaran P, Paris R, Rao P. Laser-induced thrombosis in zebrafish larvae: a novel genetic screening method for thrombosis. Methods Mol Med. 2006;129:187–95.
23 Tsai C-T, et al. In-vitro recording of adult zebrafish heart electrocardiogram – a platform for pharmacological testing. Clin Chim Acta. 2011;412:1963–7.
24 Burggren WW, Pinder AW. Ontogeny of cardiovascular and respiratory physiology in lower vertebrates. Annu Rev Physiol. 1991;53:107–35.
25 Sehnert AJ, et al. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–10.
26 Pelster B, Burggren WW. Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio). Circ Res. 1996;79:358–62.
27 Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–90.
28 Poss KD. Getting to the heart of regeneration in zebrafish. Semin Cell Dev Biol. 2007;18:36–45.
29 Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. eLife Sciences. 2015;4.
30 Chico TJA, Ingham PW, Crossman DC. Modeling cardiovascular disease in the zebrafish. Trends Cardiovasc Med. 2008;18:150–5.
31 Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–9.
32 Huisken J, Stainier DYR. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt Lett. 2007;32:2608–10.
33 Huisken J, Stainier DYR. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–75.
34 Weber M, Huisken J. Light sheet microscopy for real-time developmental biology. Current opinion in genetics & development. 2011;21:566–72.
35 Keller PJ, Stelzer EHK. Digital scanned laser light sheet fluorescence microscopy. 2010, pdb.top78 (2010).
36 Kaufmann A, Mickoleit M, Weber M, Huisken J. Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development. 2012;139:3242–7.
37 Bassi A, Schmid B, Huisken J. Optical tomography complements light sheet microscopy for in toto imaging of zebrafish development. Development. 1–19 (2015). doi:10.1242/dev.116970/-/DC1
38 Chi NC, et al. Cardiac conduction is required to preserve cardiac chamber morphology. Proc Natl Acad Sci USA. 2010;107:14662–7.
39 Hove JR, et al. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421:172–7.
40 Auman HJ, et al. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 5, e53 (2007).
41 Taylor JM, Saunter CD, Love GD, Girkin JM. Prospective gating for 3D imaging of the beating zebrafish heart in embryonic development studies. SPIE … 822716–822716–7 (2012). doi:10.1117/12.908164
42 Fahrbach FO, Voigt FF, Schmid B, Helmchen F, Huisken J. Rapid 3D light-sheet microscopy with a tunable lens. Opt Express. 2013;21:21010.
43 Dempsey WP, Fraser SE, Pantazis P. PhOTO Zebrafish: A Transgenic Resource for In Vivo Lineage Tracing during Development and Regeneration. PLoS ONE. 2012;7:e32888
44 Entcheva E. Cardiac optogenetics. AJP: Heart and Circulatory Physiology. 2013;304:H1179–H1191.
45 Bruegmann T, et al. Optogenetic control of heart muscle in vitro and in vivo. Nat Methods. 2010;7:897–900.
46 Reynaud EG, Krzic U, Greger K, Stelzer EHK. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J. 2008;2:266–75.
47 Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–9.
48 Arrenberg AB, Stainier DYR, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–4.
49 Efimov IR, Huang DT, Rendt JM, Salama G. Optical mapping of repolarization and refractoriness from intact hearts. Circulation. 1994;90:1469–80.
50 Efimov IR, Nikolski VP, Salama G. Optical imaging of the heart. Circ Res. 2004;95:21–33.
51 Gray RA, Pertsov AM, Jalife J. Spatial and temporal organization during cardiac fibrillation. Nature. 1998;392:75–8.
52 Wang Q, Shui B, Kotlikoff MI, Sondermann H. Structural basis for calcium sensing by GCaMP2. Structure. 2008;16:1817–27.
53 Kirchmaier B.C, et al. The Popeye domain containing 2 (popdc2) gene in zebrafish is required for heart and skeletal muscle development. Dev Biol. 2012;363:438–50.
54 Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–5.
55 Kralj JM, Hochbaum DR, Douglass AD, Cohen AE. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science. 2011;333:345–8.
56 Tsutsui H, Tsutsui H, Higashijima S-I, Miyawaki A, Okamura Y. Visualizing voltage dynamics in zebrafish heart. J Physiol (Lond). 2010;588:2017–21.
57 Hou JH, Kralj JM, Douglass AD. Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific developmental transitions in ionic currents. Front Physiol. (2014).
58 Panáková D, Werdich AA, Macrae CA. Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca(2+) channel. Nature. 2010;466:874–8.
59 Sedmera DD, et al. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am J Physiol Heart Circ Physiol. 2003;284:H1152–H1160.
60 Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–81.
61 Trivedi V, et al. Dynamic structure and protein expression of the live embryonic heart captured by 2-photon light sheet microscopy and retrospective registration. Biomed Opt Express. 2015;6:2056–11.
62 Li D, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500–aab3500.
63 Jorand R, et al. Deep and Clear Optical Imaging of Thick Inhomogeneous Samples. PLoS ONE. 2012;7:e35795.
64 Schmid B, et al. High-speed panoramic light-sheet microscopy reveals global endodermal cell dynamics. Nat Commun. 2013;4:2207.
65 White RM, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. 2008;2:183–9.
66 Isogai S, Horiguchi M, Weinstein BM. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Develop Biol. 2001;230:278–301.




