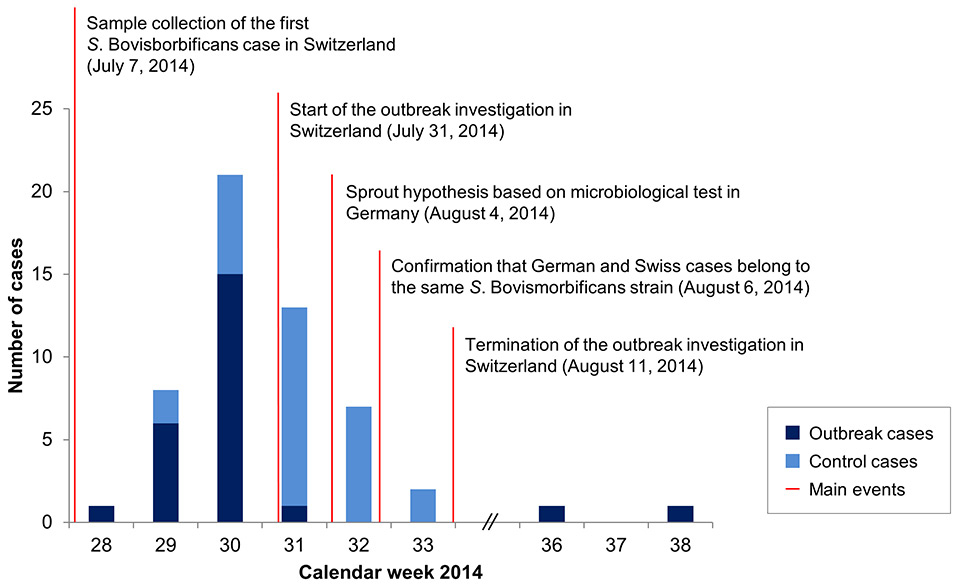
Figure 1
Epidemiological profile and main investigation events for the Salmonella Bovismorbificans outbreak, July–September 2014, Switzerland.
DOI: https://doi.org/10.4414/smw.2015.14182
Salmonella, a gram negative bacterium, is a leading cause of food-borne infections and causes outbreaks worldwide, including Switzerland [1, 2]. Salmonellaare classified into two species (S. bongori and S. enterica) and sixS. enterica sub-species; over 2 600 serovars are known [3, 4]. In Switzerland, laboratory-confirmed cases of human salmonellosis have to be reported mandatorily to the Federal Office of Public Health (FOPH) [5]. In 2013, a total of 1 271 human Salmonella cases were reported in Switzerland (15.7 per 100 000 habitants) [1]. The most frequently isolated serovars were S. Enteritidis, S. Typhimurium and S. 4,[5],12:i:- accounting for 28%, and 16% each, of total cases, respectively [1]. All serovars other than Salmonella enterica Enteritidis, or serovars that cannot be identified by the laboratories, are routinely sent to the National Reference Laboratory for Enteropathogenic Bacteria and Listeria (NENT) for serovar identification. In the case of a suspected outbreak, the NENT applies more refined typing methods such as pulsed-field gel electrophoresis (PFGE) for genotyping and clustering of the strains.
S. enterica ssp. enterica serovar Bovismorbificans (hereafter S. Bovismorbificans) previously caused outbreaks associated with lettuce in Australia (2001), raw minced pork in Germany (2005), sprouted alfalfa seeds in Finland (2009) and hummus and tahini in the United States of America (2011) [6–9]. In Switzerland, on average, fewer than 10 cases due to S. Bovismorbificans are reported annually [10]. From January to June 2014, two cases with this serovar have been reported, and in July 2014, 22 cases of S. Bovismorbificans were recorded in calendar weeks 28–31, belonging to the same PFGE cluster, indicating an outbreak event. In total, 25 outbreak cases were recorded until week 38.
Here, we report an outbreak of S. Bovismorbificans in Germany and Switzerland and its associated investigation in Swiss cases based on (i) a telephone questionnaire approach; (ii) PFGE genotyping of potential S. Bovismorbificans strains isolated from human clinical samples; and (iii) microbiological analysis of foods conducted in Germany. The primary goal of the outbreak investigation was to rapidly identify and eliminate the contamination source in order to prevent new cases. The findings, investigation approaches, timelines and the cross-border collaboration are discussed here.
In the case-case study, we included Salmonella cases that were confirmed to be caused by S. Bovismorbificans and Salmonella cases which, at the time of first reporting, were assigned to S. enterica with unconfirmed subspecies or S. entericasubsp. enterica with unconfirmed serovar (i.e. potential S. Bovismorbificans cases). Salmonella cases which were, at the time of first reporting, confirmed to be of a different species, subspecies or serovar, were excluded.
Once genotyping was completed, patients were definitively classified as outbreak cases (infected with the S. Bovismorbificans genotype specific to the outbreak) or control cases (infected with Salmonella other than the S. Bovismorbificans outbreak strain).
The outbreak investigation of Swiss cases, starting on July 31st, was based on cases notified to the FOPH within the mandatory notification system and included all cases since the identification of the first outbreak case on July 7th (sampling date) up until August 11th, 2014.
The FOPH mandated the Swiss Tropical and Public Health Institute (Swiss TPH) with the epidemiological outbreak investigation among Swiss cases. In a first step, involved laboratories and doctors were informed about the outbreak investigation and patient contact details were obtained. In a second step, patients were sent an information letter by mail. Third, patients were contacted by telephone, with a maximum of eight attempts at different times of the day, over several days. Following a successful attempt and verbal consent to participate, the patients were interviewed immediately or at the soonest convenient time. The structured questionnaire collected information on: (i) personal data for identification; (ii) questions related to symptoms and disease progression; (iii) open questions on activities and places visited in the 3 days prior to symptom onset; and (iv) prompted questions on consumption of foods, including place of purchase or consumption, in the 3 days prior to symptom onset, comprising all foods known to be possible carriers of Salmonella [11, 12]. Questions were structured in “food groups” (e.g. “fruits and berries”), sub-groups (e.g. “fruits”), and finally individual foods (e.g. “apples”), with respective skip patterns. Answer categories for consumption were “yes”, “no”, “don’t know” and “never”, where “no” was with respect to the 3 days before symptom onset and “never” meant long-term nonconsumption of a product.
The interview was carried out with the patient or, in the case of a minor, with a parent or with the patient after parental consent. Seven interviewers (five German-, one Italian- and one French-speaking) with a background in epidemiology were trained in interview techniques and on the content of the questionnaire prior to commencement of data collection. The data collection was scheduled for an indefinite period of time, until resolution of the outbreak source or a decrease of registered cases was achieved.
Data were double-entered into EpiData version 3.1 (EpiData Association; Odense, Denmark) and analysed using STATA version 13 (Stata Corp LP; College Station, USA). Univariate logistic regression was performed to calculate odds ratios (ORs) and 95% confidence intervals (CI) for comparison between cases and controls. Furthermore, χ2-test or, where applicable, Fisher’s exact test of proportions were performed to obtain p-values, where p <0.05 was considered significant.
The NENT conducted the analyses of Swiss clinical samples. Genotyping using the PulseNet harmonised PFGE protocol was performed essentially as described previously by Miller et al. (2014) [13]. XbaI digested total DNA of Salmonella Braenderup strain H9812 (ATCC BAA 664) was used as a size standard. Gel images were evaluated using BioNumerics, version 5.1 (Applied Maths; Sint-Martens-Latem, Belgium), and compared with similarity clustering using the unweighted-pair group matching algorithm and the Dice correlation coefficient with a tolerance of 1.5% and an optimisation of 1.0% [14]. The PFGE patterns were designated arbitrarily by numbering if they differed by more than four fragments; letters (a, b, c) in addition to the numbers were used to designate closely related patterns differing by only one or two fragments.
Identification of S. Bovismorbificans on food samples was confirmed through serotyping according to the White-Kauffmann-Le Minor scheme by slide agglutination with O and H antigen-specific sera (Sifin Diagnostics; Berlin, Germany) [15]. Phage typing was done according to Liesegang et al. (2002) [16]. All microbiological analyses of suspect food samples were conducted by the local health authorities of Baden-Württemberg. The strains isolated from food stuffs were serotyped at the Robert Koch Institute, Germany.
Between July 7th and July 28th 2014 (calendar weeks 28‒31, according to sampling date) the FOPH recorded 22 human cases of S. Bovismorbificans infection (see fig. 1). Cases were noted in several cantons of Switzerland, predominantly in the eastern cantons of Thurgau (n = 14) and St. Gallen (n = 4). The suspected outbreak prompted an epidemiological investigation for rapid source identification, which started on July 31st. On August 4th, the investigators were informed that in the Swiss-bordering federal state of Baden-Württemberg, Germany, a similar outbreak of S. Bovismorbificans was observed in July 2014. A total of 49 cases of S. Bovismorbificans were reported in Baden-Württemberg between calendar weeks 29–34, 2014 [17]. An association between Swiss and German cases was suspected because of the geographic proximity of the areas where cases occurred, which was taken into account by asking patients in the telephone questionnaire interview about visits to Germany. Microbiological analysis performed in Germany was able to isolate the German outbreak strain on products X and Y, two types of sprouts. However, PFGE pattern comparisons between clinical and food samples and between Swiss and German cases were still missing at this point.

Figure 1
Epidemiological profile and main investigation events for the Salmonella Bovismorbificans outbreak, July–September 2014, Switzerland.
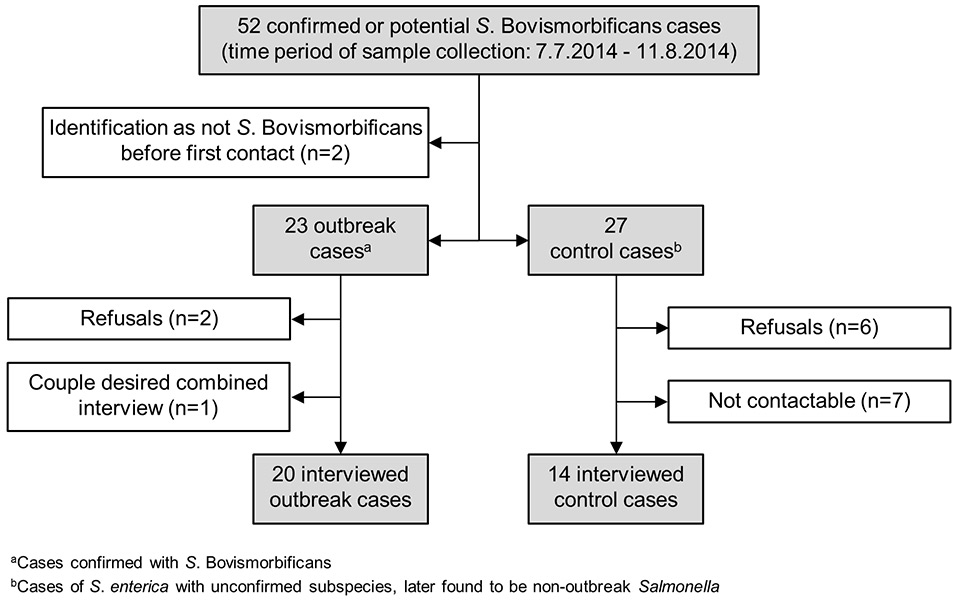
Figure 2
Participation in the Salmonella Bovismorbificans outbreak investigation, July–August 2014, Switzerland.
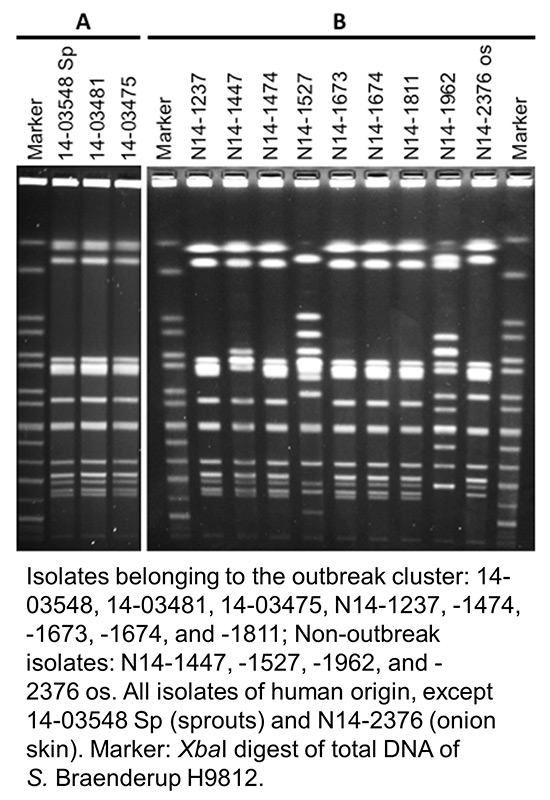
Figure 3
Pulsed-field gel electrophoresis (PFGE) profiles of selected Salmonella Bovismorbificans isolates from Germany (A) and Switzerland (B).
On August 6th, telephone interviews started in Switzerland and, at the same time, the NENT confirmed that the S. Bovismorbificans cases in Switzerland and Germany belonged to the same strain, suggesting a common source. No new outbreak cases were observed in Switzerland during calendar weeks 32 and 33, which caused the FOPH to terminate the outbreak investigation by August 11th. After the outbreak investigation, two further outbreak cases were registered in Switzerland in calendar weeks 36 and 38. Routine surveillance of Salmonella cases in Switzerland did not detect any outbreak cases after week 38 until the end of 2014.
The most common self-reported symptoms experienced by outbreak cases were diarrhoea (95.0% vs 92.9% in controls), crampy abdominal pain (65.0% vs 50.0% in controls) and fever (80.0% vs 35.7% in controls). The symptoms in outbreak cases lasted on average 9.1 ± 6.9 standard deviation (SD) days (9.5 ± 7.4 SD in controls), and 6 outbreak cases (30.0%) were hospitalised for this disease episode (50.0% in controls).
During the investigation period lasting from July 7th to August 11th 2014, according to sampling date, a total of 52 confirmed or potential S. Bovismorbificans cases were recorded (fig. 2). Among those, two patients were excluded as the serovar was identified as non-Bovismorbificans before the first telephone contact. Thirty-four of the remaining 50 cases could be interviewed (20 outbreak and 14 control cases). With seven patients no contact could be established, eight patients refused to participate and one married couple, both outbreak salmonellosis patients, participated in the interview together as they had had very similar food habits in the 3 days prior to onset of symptoms. The two outbreak cases registered after termination of the outbreak investigation on August 11th 2014 and were not further investigated.
Table 1 shows “foods” and “food groups” consumed by outbreak and control cases in the 3 days prior to symptom onset. Similarities of food consumption in outbreak cases showed that “sprouts and salads (all sorts)” and “salads (all sorts)” were most frequently consumed, by 19 of 20 outbreak cases. In comparison, products from these food groups were each consumed by 8 of 14 control cases. Differences between outbreak and control cases were assessed by use of univariate analysis with all “foods” and “food groups” and ranked according to their OR. “Sprouts and salads (all sorts)” and “salads (all sorts)” had the highest ORs (both 14.3, 95% CI 1.47–138.27). This was followed by “sprouts (all sorts)” and “cereals (all sorts)” each with an OR of 10.6 (95% CI 1.16–97.59). One outbreak case reported never eating “cereals”.
Investigations on activities and places visited, for example visits to restaurants or similar establishments, in the 3 days prior to symptom onset revealed that:
1. 100% of outbreak cases reported having eaten in a restaurant at least once;
2. 80% of outbreak cases had visited places in Baden-Württemberg;
3. 100% of the outbreak cases who had visited Baden-Württemberg had eaten either “sprouts” or “salads” or a combination thereof in local restaurants;
4. together, the outbreak cases consumed in total 47 times the suspected food group “sprouts and salads” from the following outlets: 24 times in restaurants, 13 times purchased from retailers, 10 times from “unknown or other location”;
5. no control case reported a visit to Baden-Württemberg; and
6. together, the control cases consumed in total 28 times the suspected food group “sprouts and salads” from the following outlets: once in a restaurant, 17 times purchased from retailers, 10 times from “unknown or other location”.
Genotyping of human clinical isolates revealed that one single clone was responsible for the outbreak and that all outbreak cluster isolates (n = 38 in Germany; n = 25 in Switzerland) shared an indistinguishable banding pattern independent of the country of origin (see fig. 3A for German and fig. 3B for Swiss isolates). The discriminatory power of the method was high, since profiles representing nonoutbreak isolates of S. Bovismorbificans were clearly distinguishable (e.g. see lanes “N14-1447”, “-1527”, “-1962” and “-2376os” in fig. 3B). PFGE profiles of all isolates reported to belong to the outbreak cluster were indistinguishable from the outbreak profile.
In Germany, two sprout products (X and Y) were identified by means of microbiological analysis. Salmonella strains found on both products were sent for genotyping. The two sprout strains were at the same time compared with a random selection of five human outbreak isolates originating from Baden-Württemberg (see line 14-03548 Sp in fig. 3A representing the sprout isolate). All strains belonged to the same PFGE type.
| Table 1:Consumption and prioritisation of risk ‘foods’ and ‘food groups’ consumed in the 3 days prior to illness according to the odds ratio. | ||||||
| Food consumption | Outbreak cases (n = 20) | Control cases (n = 14) | Odds ratio (95% confidence interval) | p-value | ||
| Yes (%) | No (%) | Yes (%) | No (%) | |||
| Sprouts and salads (all sorts) | 19 (95.0) | 1 (5.0) | 8 (57.1) | 6 (42.9) | 14.3 (1.47–138) | 0.01 |
| Salads (all sorts) | 19 (95.0) | 1 (5.0) | 8 (57.1) | 6 (42.9) | 14.3 (1.47–138) | 0.01 |
| Sprouts (all sorts) | 9 (45.0) | 11 (55.0) | 1 (7.1) | 13 (92.9) | 10.6 (1.16–97.6) | 0.02 |
| Cereals (all sorts) | 9 (45.0) | 11 (55.0) | 1 (7.1) | 13 (92.9) | 10.6 (1.16–97.6) | 0.02 |
| Pepper | 7 (35.0) | 12 (65.0) | 1 (7.1) | 13 (92.9) | 7.0 (0.75–65.2) | 0.10 |
| Cucumber | 10 50.0) | 10 (50.0) | 2 (14.3) | 12 (85.7) | 6.0 (1.06–34.0) | 0.06 |
| Eggs (all preparation variants) | 12 (60.0) | 7 (40.0) | 3 (21.4) | 11 (78.6) | 5.5 (1.16–26.1) | 0.04 |
In an S. Bovismorbificans outbreak in July 2014 in Switzerland, 20 outbreak cases and 14 control cases were investigated in a case-case study by means of a telephone questionnaire interview. The questionnaire tool was able to narrow down the contamination source with clear indications of (i) a statistically significant association between the consumption of “sprouts and salads” and an S. Bovismorbificans infection; and (ii) the consumption of the suspected “sprouts and salads” in restaurants in Baden-Württemberg, Germany.
PFGE genotyping of clinical isolates in Germany and Switzerland were compared with food isolates of two types of sprouts genotyped in Germany. A common outbreak strain could be identified in all outbreak samples and was responsible for 49 and 25 cases in Germany and Switzerland, respectively, between calendar weeks 28 and 38.
The characteristation of the outbreak, i.e. its contamination source and place of consumption of the high risk product, had its perils. Meal ingredients in restaurants can be challenging for consumers to know and recall. In our investigation, “salads” showed a stronger association with outbreak cases than “sprouts”. It is likely that outbreak cases who did not mention the consumption of sprouts consumed these unknowingly as a side-dish or garnish in restaurant meals. In fact, previous Salmonella outbreaks associated with sprouts have shown low self-reported sprout consumption [18–21]. For outbreak cases without reported visits to Baden-Württemberg (n = 4; 20%), the origin of infection remains largely unexplained. However, one common factor among the outbreak cases who did not visit Germany was that they all ate on at least one occasion in a restaurant in Switzerland. According to the manufacturer, products X and Y were not delivered to Switzerland, but there is a possibility that the products were purchased at a wholesale store in Germany and processed in Swiss restaurants [22]. For the two cases that were registered only after the outbreak investigation, no further inquiries have been made. Origin of infection and especially the late case recognition are difficult to elucidate. Salmonella can be detected in stools up to 1 month after infection [23]. Hence, it is possible that these patients showed no acute symptoms and stool samples were taken for another episode of illness. However, it also remains unclear whether these two patients visited Germany or consumed a residual of product X or Y.
PFGE typing of isolates and microbiological analysis for clinical and food samples are standard methods [24], but approaches used for patient surveys are more diverse. Questionnaire tools are often used for epidemiological outbreak investigations, although in different forms (self-administered vs interview, open- vs closed-ended), at different entry points (face-to-face interview vs telephone interviews) or at different times (at sample collection vs after diagnosis) [25–28]. The questionnaire used in this survey, containing mainly closed-ended questions, was derived from the so-called “shotgun” questionnaire put forward as an outbreak investigation tool for hypothesis generation [29]. This trawling questionnaire was adapted to the Swiss context and pre-tested with cases (having gastric symptoms) vs controls (not having gastric symptoms) in early 2014 [30]. At the start of this investigation, it was not confirmed by PFGE analysis that the Swiss and German cases originated from the same outbreak strain, and a broad inquiry covering all potential contamination sources was therefore indicated [31]. The microbiological findings during the investigation transformed it from an exploratory investigation without a source hypothesis into a hypothesis-driven investigation. Previous studies have shown that use of initial trawling questionnaire interviews with a small number of cases (e.g. 10 cases) to generate a hypothesis followed by hypothesis-testing in the entire study population can be a cost-effective approach [32]. In addition, such fast hypothesis generation can present a formidable base for rapid onset of focused microbiological food analysis.
Patients with other Salmonella infections were considered suitable control cases in this investigation since cases and controls are assumed to have similar risk factors. As in other studies, our investigation was able to show significant differences between outbreak and nonoutbreak Salmonella cases [21]. An additional advantage of using nonoutbreak cases as controls is that recruitment is faster and acceptance for participation is likely elevated compared to nonaffected, population-matched controls [20].
Our investigation highlights the importance of cross-country collaboration for a timely identification of the source and prevention of an expanded outbreak. On July 29th 2014, the outbreak was detected by the Swiss mandatory surveillance system. On July 31st 2014, the FOPH informed all Swiss cantonal physicians about the outbreak. In response, the cantonal physician of Thurgau signaled a potential association with an ongoing Salmonella outbreak in Baden-Württemberg. The FOPH immediately contacted the German health authorities, which provided important details (e.g. location of cases, Salmonella strain) that helped to focus the outbreak investigation in Switzerland. The incidence of Swiss outbreak cases decreased strongly before the outbreak strain was confirmed on sprout products X and Y, suggesting that (i) the measures taken in German restaurants influenced the observed decline; (ii) the source was eliminated without specific interventions; or (iii) the contaminated products were used up. Except for the two late cases, no new cases were registered after source elimination, and hence, there was no need for either inspections in restaurants, recall of food products or public alerts in Switzerland.
The time period between sample collection and the telephone questionnaire interview was relatively long (i.e. 19.8 ± 7.0 SD days), introducing a recall bias into the data. This was due to (i) naturally delayed onset of the outbreak investigation; (ii) time elapsing between patient tracing and contact attempts; and (iii) frequent absence of patients due to the holiday season. Therefore, the patients could not always remember precisely their food consumption in the relevant period. The sample size for both cases and controls was small, causing wide CIs around the estimates. After obtaining the results from the microbiological testing of foods in Germany, interviewers were sensitised for more specifically inquiring about stays, activities and food consumption patterns abroad.
The telephone-based questionnaire tool used in the frame of a case-case study is considered appropriate to deliver precise indications on vehicle and location of the source of food-borne infections in a rapid manner. In this investigation, a specific product could not be identified by questionnaire interviews alone, possibly owing to the nature of the outbreak, i.e. its contamination source and place of consumption. However, the combination of approaches proved successful, and the microbiological testing was a crucial determinant in identification of the contaminated products X and Y. Recurring outbreaks associated with sprouts suggest that they are an increasingly frequent carrier of various Salmonellaspecies [8, 18, 19, 21, 33, 34]. Susceptible persons, e.g. immunosuppressed people or people having other debilitating conditions, should be careful about consuming raw sprouts and other risky products (e.g. raw eggs).
1 European Food Safety Authority (EFSA). Zoonoses monitoring: Switzerland: Trends and sources of zoonoses and zoonotic agents in humans, foodstuffs, animals and feedingsstuffs. 2013; Parma: EFSA.
2 Schmid H, Baumgartner A. Epidemiology of infections with enteric salmonellae in Switzerland with particular consideration of travelling activities. Swiss Med Wkly. 2013;143:w13842.
3 Tindall BJ, Grimont PA, Garrity GM, Euzeby JP. Nomenclature and taxonomy of the genus Salmonella. Int J Syst Evol Microbiol. 2005;55(Pt 1):521–4.
4 Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, et al. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2010;161(1):26–9.
5 Bundesrecht. Verordnung über die Meldung übertragbarer Krankheiten des Menschen. 1999; Bern: Schweizerische Eidgenossenschaft.
6 Stafford RJ, McCall BJ, Neill AS, Leon DS, Dorricott GJ, Towner CD, et al. A statewide outbreak of Salmonella Bovismorbificans phage type 32 infection in Queensland. Commun Dis Intell Q Rep. 2002;26(4):568–73.
7 Gilsdorf A, Jansen A, Alpers K, Dieckmann H, van Treeck U, Hauri AM, et al. A nationwide outbreak of SalmonellaBovismorbificans PT24, Germany, December 2004–March 2005. Euro Surveill. 2005;10(3):pii=2667.
8 Rimhanen-Finne R, Niskanen T, Lienemann T, Johansson T, Sjoman M, Korhonen T, et al. A nationwide outbreak of Salmonella Bovismorbificans associated with sprouted Alfalfa seeds in Finland, 2009. Zoonoses Public Hlth. 2011;58(8):589–96.
9 Centers for Disease Control and Prevention (CDC). Multistate outbreak of Salmonella serotype Bovismorbificans infections associated with hummus and tahini - United States, 2011. MMWR Morb Mortal Wkly Rep. 2012;61(46):944–7.
10 National Reference Laboratory for Enteropathogenic Bacteria and Listeria (NENT), Bundesamt für Gesundheit (BAG). EPIS Alert NENT BAG 060814. 2014; Bern: NENT, BAG.
11 Heymann DL. Control of communicable diseases manual: 19th edition. 2008. Washington DC: American Public Health Association.
12 Bundesamt für Gesundheit (BAG). Lebensmittelbedingte Gruppenerkrankungen in der Schweiz. 2012; Bern: BAG.
13 Miller T, Braun PG, Fehlhaber K, Prager R, Pfeifer Y, Rabsch W. Typing of Salmonella enterica serovar Infantis isolates from 51 outbreaks in Germany between 1974 and 2009 by a novel phage-typing scheme. Epidemiol Infect. 2014;142(1):75–83.
14 Centers for Disease Control and Prevention (CDC). Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia colinon-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. 2013; Atlanta: Centers of Disease Control.
15 Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars: 9th edition. 2007; Geneva: World Health Organization Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur.
16 Liesegang A, Davos D, Balzer JC, Rabsch W, Prager R, Lightfoot D, et al. Phage typing and PFGE pattern analysis as tools for epidemiological surveillance of Salmonella enterica serovar Bovismorbificans infections. Epidemiol Infect. 2002;128(2):119–30.
17 Landesgesundheitsamt Baden-Württemberg. Infektionsbericht Baden-Württemberg: Meldewoche 34. 2014; Stuttgart: Landesgesundheitsamt Baden-Württemberg.
18 Van Beneden CA, Keene WE, Strang RA, Werker DH, King AS, Mahon B, et al. Multinational outbreak of Salmonella enterica serotype Newport infections due to contaminated Alfalfa sprouts. JAMA. 1999;281(2):158–62.
19 Buchholz U, Bernard H, Werber D, Bohmer MM, Remschmidt C, Wilking H, et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med. 2011;365(19):1763–70.
20 Gobin M, Launders N, Lane C, Kafatos G, Adak B. National outbreak of Salmonella Java phage type 3b variant 9 infection using parallel case-control and case-case study designs, United Kingdom, July to October 2010. Euro Surveill. 2011;16(47):pii=20023.
21 Bayer C, Bernard H, Prager R, Rabsch W, Hiller P, Malorny B, et al. An outbreak of Salmonella Newport associated with mung bean sprouts in Germany and the Netherlands, October to November 2011. Euro Surveill. 2014;19(1):pii=20665.
22 Bundesamt für Lebensmittelsicherheit und Veterinärwesen (BLV), Schnellwarnsystem für Lebens- und Futtermittel (RASFF). Lebensmittelüberwachung: Gutachten zu Sprossen. 2014; Bern: BLV, RASFF.
23 Robert Koch Institute (RKI). Salmonellose (Salmonellen-Gastroenteritis): RKI-Ratgeber für Ärzte Berlin: RKI; 2011 [cited January 2015]. Available from: http://www.rki.de/DE/Content/Infekt/EpidBull/Merkblaetter/Ratgeber_Salmonellose.html.
24 Refsum T, Heir E, Kapperud G, Vardund T, Holstad G. Molecular epidemiology of Salmonella enterica serovar Typhimurium isolates determined by pulsed-field gel electrophoresis: comparison of isolates from avian wildlife, domestic animals, and the environment in Norway. Appl Environ Microbiol. 2002;68(11):5600–6.
25 Ghosh TS, Patnaik JL, Alden NB, Vogt RL. Internet- versus telephone-based local outbreak investigations. Emerg Infect Dis. 2008;14(6):975–7.
26 Whelan J, Noel H, Friesema I, Hofhuis A, de Jager CM, Heck M, et al. National outbreak of Salmonella Typhimurium (Dutch) phage-type 132 in the Netherlands, October to December 2009. Euro Surveill. 2010;15(44):pii=19705.
27 Raguenaud ME, Le Hello S, Salah S, Weill FX, Brisabois A, Delmas G, et al. Epidemiological and microbiological investigation of a large outbreak of monophasic Salmonella Typhimurium 4,5,12:i:- in schools associated with imported beef in Poitiers, France, October 2010. Euro Surveill. 2012;17(40):pii=20289.
28 Kunwar R, Singh H, Mangla V, Hiremath R. Outbreak investigation: Salmonella food poisoning. Med J Armed Forces India. 2013;69(4):388–91.
29 Oregon Health Authority. Shotgun hypothesis-generating questionnaire. 2014; Portland: Oregon Health Authority.
30 Kiefer S, Künzli E, Hatz C. Kompetenzzentrum für epidemiologische Ausbruchsuntersuchungen: Prätest Bericht. 2013; Basel, Zürich: Swiss Tropical and Public Health Institute, Institut für Sozial- und Präventivmedizin der Universität Zürich.
31 Hächler H, Marti G, Giannini P, Lehner A, Jost M, Beck J, et al. Outbreak of listerosis due to imported cooked ham, Switzerland 2011. Euro Surveill. 2013;18(18):pii=20469.
32 Inns T, Beasley G, Lane C, Hopps V, Peters T, Pathak K, et al. Outbreak of Salmonella enterica Goldcoast infection associated with whelk consumption, England, June to October 2013. Euro Surveill. 2013;18(49):pii=20654.
33 Stewart DS, Reineke KF, Ulaszek JM, Tortorello ML. Growth of Salmonelladuring sprouting of alfalfa seeds associated with salmonellosis outbreaks. J Food Prot. 2001;64(5):618–22.
34 Safranek T, Leschinsky D, Keyser A, O'Keefe A, Timmons T, Holmes S, et al. Outbreak of Salmonella serotype Saintpaul infections associated with eating alfalfa sprouts – United States, 2009. MMWR Morb Mortal Wkly Rep. 2009;58(18):500–3.
Diclosure statement: No financial support and no other potential conflict of interest relevant to this article was reported.