Fish β-parvalbumin acquires allergenic properties by amyloid assembly
DOI: https://doi.org/10.4414/smw.2015.14128
Javier
Martínez, Rosa
Sánchez, Milagros
Castellanos, Ana M
Fernández-Escamilla, Sonia
Vazquez-Cortés, Montserrat
Fernández-Rivas, Maria
Gasset
Summary
PRINCIPLES: Amyloids are highly cross-β-sheet-rich aggregated states that confer protease resistance, membrane activity and multivalence properties to proteins, all essential features for the undesired preservation of food proteins transiting the gastrointestinal tract and causing type I allergy.
METHODS: Amyloid propensity of β-parvalbumin, the major fish allergen, was theoretically analysed and assayed under gastrointestinal-relevant conditions using the binding of thioflavin T, the formation of sodium dodecyl sulphate- (SDS-) resistant aggregates, circular dichroism spectroscopy and atomic force microscopy fibril imaging. Impact of amyloid aggregates on allergenicity was assessed with dot blot.
RESULTS: Sequences of β-parvalbumin from species with commercial value contain several adhesive hexapeptides capable of driving amyloid formation. Using Atlantic cod β-parvalbumin (rGad m 1) displaying high IgE cross-reactivity, we found that formation of amyloid fibres under simulated gastrointestinal conditions accounts for the resistance to acid and neutral proteases, for the presence of membrane active species under gastrointestinal relevant conditions and for the IgE-recognition in the sera of allergic patients. Incorporation of the anti-amyloid compound epigallocatechin gallate prevents rGad m 1 fibrillation, facilitates its protease digestion and impairs its recognition by IgE.
CONCLUSIONS: the formation of amyloid by rGad m 1 explains its degradation resistance, its facilitated passage across the intestinal epithelial barrier and its epitope architecture as allergen.
Introduction
More than 5% of the population suffer from type I food allergy, an immunoglobulin E- (IgE-) mediated hypersensitivity disease provoked by food proteins [1–3]. Type I food allergy involves two main phases; a sensitisation step and an effector phase [3–5]. The sensitisation occurs in the gastrointestinal tract (GIT) after the contact with the ingested allergen and consists in a series of events leading to an overproduction of allergen-specific IgE able to bind to the high-affinity IgE receptor FcεRI on the surface of basophils and mast cells. In the effector phase, the causative allergen crosses the intestinal epithelium and cross-links the IgE-FcεRI complexes, provoking effector cell activation and the release of allergy mediators. Although individual reactivity depends on genetic and environmental factors, stability of food allergens through the GIT is an essential requirement for their pathogenicity [3, 6–9]. The stability confers resistance to heat treatments, to drastic pH changes (pH 2–7.5), to acid and neutral proteases and to detergents, and permits the preservation of immunogenic motifs for interaction with the epithelial immune system or passage through the epithelial barrier in order to induce sensitisation and systemic symptoms [8, 9]. These properties are shared by more than 120 molecular architectures with different folds, suggesting as yet unknown common structural threats [10, 11].
Pathogenic proteins such as prions display such stability by virtue of their amyloid fibrillar state [12, 13]. Amyloid fibrils are insoluble protein aggregates with a highly compact spine based on a cross-β-sheet structure [14–18]. This structure comprises an indefinitely repeating intermolecular β-sheet motif, in which each pair of intermolecular β sheets interdigitate their side chains as a steric zipper. Amyloids are very stable and more resistant to hydrolysis than the folded globular protein, and upon the proper signal they can release monomers and create a population of different oligomeric and polymeric intermediates [18, 19]. Amyloid fibrils are considered poorly immunogenic, but their aggregated fragments as prefibrillar oligomers and fibrillar oligomers have yielded very valuable conformational antibodies [20, 21]. Therefore, assembly into amyloid-like structures by food allergens could explain some of their pathogenic properties.
Materials and methods
Chemicals and proteins
All reagents were of the highest grade commercially available. Thioflavin T was obtained from Sigma. A Chelex resin (Bio-Rad) was used to remove contaminant trace metals from all solutions. Recombinant Gad m 1 (rGad m 1) was produced from a pET15b construct containing the synthetic open reading frame sequence of Atlantic cod parvalbumin A51874 (Genscript). Protein was isolated from the soluble fraction of sonicated bacterial cells and purified by Ni2+-NTA (nickel-nitrilotriacetic acid) chromatography. Removal of His-tag was performed by a 3-h digestion with thrombin, following ultrafiltration through a 30 kDa cutoff filter. Filtrates containing rGad m 1 were extensively dialysed against mQ H20 and then lyophilised. Protein was quantified using Bio-Rad protein assay. Before use, rGad m 1 was equilibrated by dialysis in 5 mM Hepes pH 7.4 and centrifuge at 12,000 × g for 15 min at 4 °C to remove any insoluble material. Fibrils from Aβ1–40 and HaPrP23–231 were formed as described [22, 23].
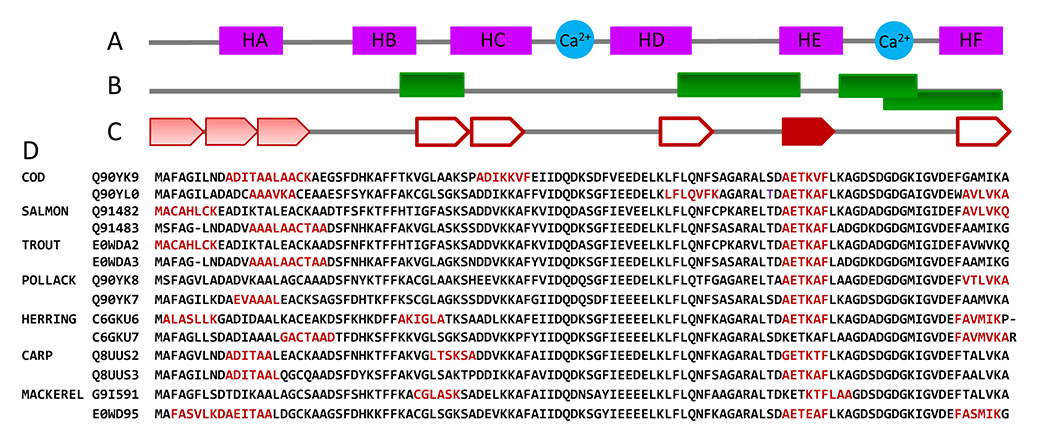
Figure 1
Propensity of fish β-parvalbumin sequences to form amyloids. (A) Fish β-parvalbumin fold described for cod (PDB ID:2MBX), carp (PDB ID: 4CPV), hake (PDB ID: 1BU3), pike (PDB ID: 1PVB) and whiting (PDB ID: 1A75) chains is largely conserved and consists of six α-helices pairing as AB, CD and EF and forming three EF-hand motifs, two of which (CD and EF) coordinate Ca2+. (B) Immunological reactive sites described for fish β-parvalbumins, shown as green rectangles, embrace the spacers between motifs and on the C-terminal motif [29–31]. (C) Some hexapeptides with adhesive properties found in β-parvalbumin sequences from commercially valuable fish and known allergic potential using Zipper Db flank the immunological reactive sites. Hexapeptides (single or extended) are depicted in red, by (C) arrows (the saturation of the colour is proportional to the similarity of the segments) and by (D) their sequences.
Human sera
Fish allergic patients had convincing case histories of fish allergy, positive skin prick tests to codfish (≥5 mm mean wheal diameter), and serum specific IgE to cod (30.3 and 9.6 kU/l) and to rGad c 1 (14.2 and 18.5 ku/l, respectively) and a positive double-blind placebo-controlled food challenge with codfish (ImmunoCAP, ThermoFisher Scientific, Uppsala, Sweden). Sera were stored at –20 °C until use. Written informed consent was obtained from patients and the study was approved by the Ethics Committee of the Hospital Clínico San Carlos (Madrid).
Amyloidogenic propensity analysis
The amyloidogenic propensity of fish β-parvalbumins was analysed using the Zipper Db algorithm [17]. The sequences considered were: Atlantic cod (Gadus morhua) ‒ Q90YK9, Q90YL0, A51874; Atlantic salmon (Salmo salar) ‒ Q91482, Q91483, B5DH15, B5DH16, E0WD98, E0WD99; Rainbow trout (Salmo gairdneri, Oncorhynchusmykiss) ‒ E0WDA2, E0WDA3; Alaska pollock (Theragra chalcogramma, Gadus chalcogramma) ‒ Q90YK8, Q90YK7; Atlantic herring (Clupea harengus) ‒ C6GKU6, C6GKU7; carp (Cyprinus carpio) ‒ Q8UUS2, Q8UUS3, E0W92, E0W93; mackerel (Scomber japonicus, Trachurus japonicas, Scomber vernalis) ‒ G9I591, E0WD95, Q3C2C3, Q3C2C4, D3GME4, C0LEK8; hake (Merluccius bilinearis, Merluccius merluccius, Merluccius australis, Merluccius senegalensis) ‒ P56503, P02620, P86745, P86778.
Aggregation assays
Recombinant Gad m 1 solutions at concentrations of 0.5–5 mg/ml were prepared in simulated gastric fluid (SGF: 50 mM glycine pH 2.0, 35 mM NaCl) or simulated intestinal fluid (SIF: 50 mM Tris pH 7.5, 35 mM NaCl), in both cases supplemented with either 5 mM EDTA or 5 mM CaCl2. Simulated gastrointestinal fluid (SGIF) was achieved by a 30-min incubation in SGF followed by the addition of 1/5 (V/V) of 1.5 M Tris pH 8.0. When required, fibres were harvested by a 100,000 x g centrifugation for 1 h using an OptimaTM MAX Beckman ultracentrifuge, and the pellet and supernatant fractions used for analysis. The binding of ThT for the detection of amyloid was performed as described [22]. Detergent resistant aggregates were assayed by means of SDS-polyacrylamide gel electrophoreses (SDS–PAGE) in 17% polyacrylamide gels; the proteins were loaded without heating and visualised with either Coomassie blue or silver staining.
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy experiments were performed with a Jasco J-820 spectropolarimeter equipped with a Peltier-controlled thermostated cell holder. Far-UV CD spectra were recorded for 30 μM rGad m 1 in SGF and SIF supplemented with either 1 mM EDTA or 1 mM CaCl2. Spectral analysis was performed as described [24].
Dynamic light scattering
Dynamic light scattering (DLS) data were acquired at 25 °C by use of Wyatt Dyna-Pro DLS system with a 1-mm path length 12 μl quartz cuvette. Samples were filtered with a 0.22 μm Whatman Anodisc-13 filter. Data were collected with a 5-second acquisition time, 20 acquisitions per measurement at laser power 100% (buffer) and 85% (protein samples) and were analysed with the Dynamics software.
Atomic force microscopy
For visualisation with atomic force microscopy (AFM), 5-μl sample aliquots of 0.5 mg/ml protein concentration were absorbed to freshly cleaved mica. Images were obtained using a MultiMode Veeco microscope with 125-µm lateral range and 5-µm vertical range J-scanner and NanoScope IIIa controller. Rectangular cantilevers with tetrahedral tips for dynamic (tapping) mode in air were purchased from Olympus (OMCL-AC240TS). Software to obtain and treat the images was supplied with the instrumentation (NanoScope). For specific AFM analysis, we used the free software WSxM 4.0 develop 13 (Nanotec).
Simulated gastrointestinal digestion
Recombinant Gad m 1 was both freshly dissolved or incubated for 36 h in SGF (1–10 mg/ml protein concentration) with or without 20 μM (−)-epigallocatechin gallate (EGCG, Sigma). The mixture was maintained at 37 °C with gentle shaking, and the reaction was started with the addition of pepsin (Sigma) at a ratio 1:70 w/w enzyme:substrate. After 30 min, the digestion was stopped by increasing the pH to 7.5 with 1.5 M Tris-HCl. For simulating the intestinal digestion, the reaction products were supplemented with proteinase K (1:70 w/w, protease:substrate) and digestion was allowed for 30 min at 37 °C. The digestion was stopped by adding phenylmethylsulfonyl fluoride (PMSF) at a final concentration of 2 mM. Control experiments without proteases or with bovine serum albumin (BSA) instead of rGad m 1 were also performed. The reaction products were analysed by means of SDS-PAGE using 17% polyacrylamide gels.
Dot-blot analysis
The reactivity of protein species against conformation-dependent antibodies was evaluated in a dot-blot analysis using the anti-amyloid fibrils OC antibody (AB2286 Merck Millipore, 1/2000 dilution), the A11 anti-amyloid oligomer antibody (AB9234 Merck Millipore, 1/2000 dilution), and sera from patients allergic to cod fish (1/10 dilution). Briefly, aliquots of rGad m1 (1–50 ng) under the different treatments were spotted in triplicates on a nitrocellulose membrane and treated with and without 6 M GdnCl. Immunodetection was performed by 1 h of incubation with primary antibodies, followed by extensive washes and 30 min incubation with horseradish peroxidase-labelled antibody, either mouse monoclonal B3102E8 anti-human IgE (Abcam, diluted 1:2000) or goat anti-rabbit IgG (1:5000 diluted; Sigma). The signal was developed with the ECL-western-blotting reagent (Biorad), and detected with ChemiDoc XRS equipment [25].
Results
To address the question of whether degradation properties and preservation of monomers involves amyloid-like aggregates we chose β-parvalbumins, the main elicitors of IgE-mediated reactions in fish-allergic individuals [26]. The β-parvalbumins are Ca2+ binding proteins of about 12 kDa and contribute >2.5 mg per gram to raw fish muscle [26–28]. They fold into a structure consisting of three EF-hand motifs (AB, CD and EF) (fig. 1A), of which only CD and EF can bind divalent cations (Ca2+ and/or Mg2+) [26, 27]. The major immunologically reactive sites have been found on the junction between the motifs (regions 33–44, 65–74) and on the EF region (segments 88–96 and 95–109) [26, 29–31]. Ca2+-bound forms are extremely stable and even cooking cannot alter their allergenicity [26]. However, loss of calcium causes an altered conformation, with decreased stability and impaired IgE recognition [26, 32].
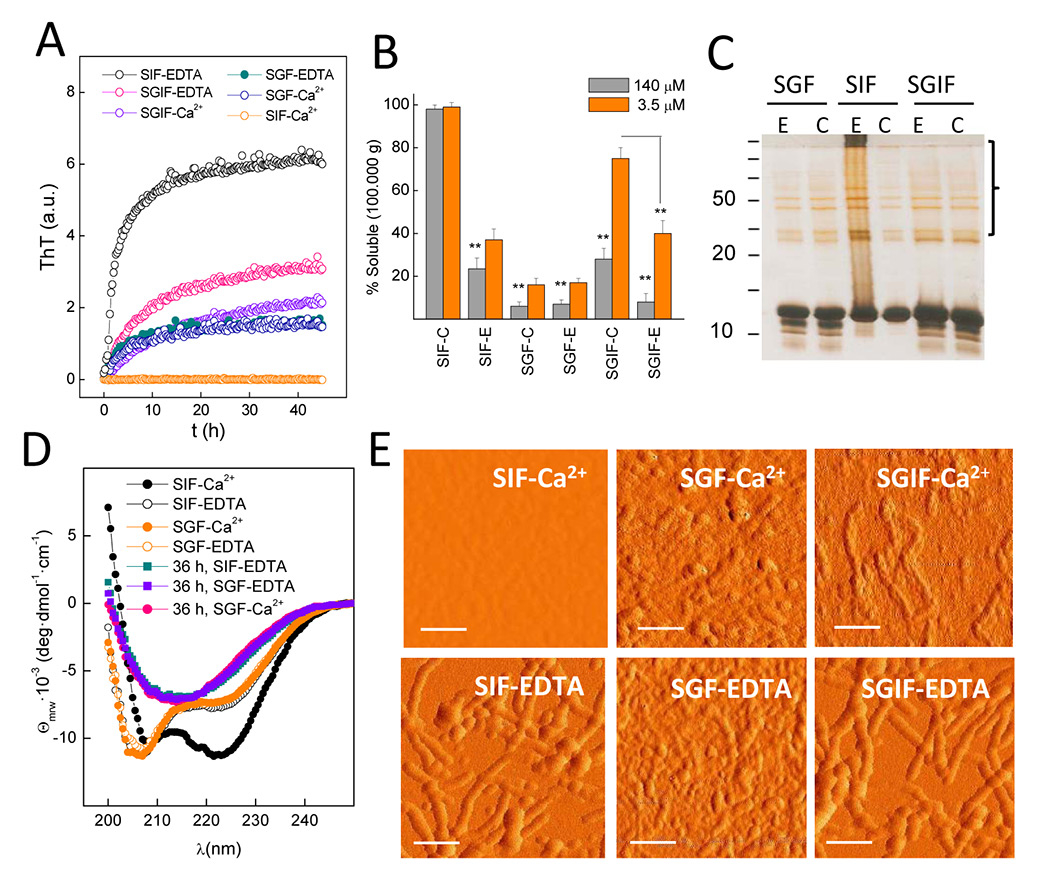
Figure 2
Amyloid-like aggregation of cod β-parvalbumin. (A) Kinetics of thioflavin T (ThT) binding indicate rGad m 1 fibril formation under simulated gastric fluid (SGF), simulated intestinal fluid (SIF) supplemented with 5 mM EDTA and simulated gastrointestinal fluid (SGIF) containing either 5 mM EDTA or 5 mM CaCl2. Reactions were performed at 37 °C using 140 μM rGad m1, and the measured fluorescence counts normalised. (B) Percentage of soluble rGad m 1 recovered in the supernatants of a 100,000 x g centrifugation after 36-h incubations in SIF, SGF and SIGF, supplemented with 5 mM EDTA (E) or 5 mM CaCl2. (C) show formation of insoluble aggregates under those conditions with increased ThT binding. Incubations were performed at 3.5 and 140 μM rGad m 1. Solubility percentages were determined by measuring the protein concentration in both the soluble and the pellet fraction using triplicates. Bar charts are presented as mean ± standard deviation (n = 3); ** p < 0.05. (C) Analysis of rGad m 1 aggregates by use of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining shows bands at high molecular weight corresponding to SDS-resistant aggregates. (D) Circular dichroism (CD) spectra of 60 μM rGad m 1 in SGF and SIF supplemented with either 5 mM EDTA or 5 mM CaCl2 indicate β-sheet structures after 36 h incubation of the apoforms. (E) Atomic force microscopy (AFM) images of rGad m1 incubated for 36 h under SGF, SIF and SGIF supplemented with 5 mM EDTA or 5 mM CaCl2 indicating the formation of aggregates and of their fibrillar shape. Scale bars 100 nm.
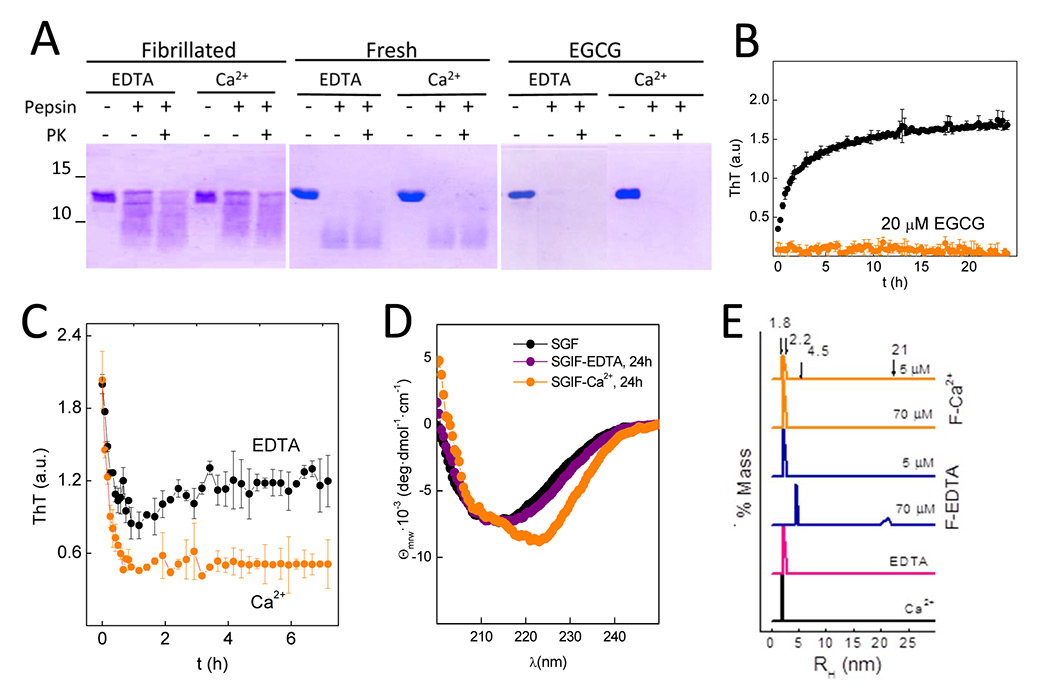
Figure 3
Amyloid fibrils protect rGad m1 from protease digestion and function as depots releasing distinct species.(A) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of gastric (pepsin at pH 2.0) and gastrointestinal (pepsin at pH 2.0 followed by proteinase K at pH 7.4) digestions of freshly prepared solutions or after 36 h of fibrillation in SGF, in the absence and presence of 20 μM of epigallocatechin gallate (ECGC), show amyloid-assembly protects monomers from proteolysis. Gels were stained with Coomassie blue. (B) Incubation of rGad m 1 with ECGC (20 μM) prevents the increase in thioflavin T (ThT) fluorescence due to fibrillation. Incubation was performed for 15 min in simulated gastric fluid (SGF) and then brought to simulated intestinal fluid (SIF) conditions. Data correspond to two independent experiments performed in triplicate. (C) Time-course decrease of ThT fluorescence of rGad m 1 fibrils resuspended in SIF containing either 5 mM EDTA or 5 mM CaCl2 indicates amyloid disassembly. (D) Evolution of the circular dichroism (CD) spectrum of rGad m1 fibrils after resuspension in SIF containing either 5 mM EDTA or 5 mM CaCl2 reveal effects of amyloid disassembly on secondary structure. Spectra were recorded 3 h after resuspension. (E) DLS analysis of soluble rGad m 1 obtained in the supernatant of a 100,000 x g centrifugation of amyloid fibrils resuspended in SGF and SIF containing either 5 mM EDTA or 5 mM CaCl2 show the release of monomers and oligomers. Species with RH of 1.8 ± 0.1 nm and 2.2 ± 0.2 nm correspond to Ca2+-bound and free monomers, and those with 4.5 ± 0.5 nm and 21 ± 0.5 nm to oligomers.
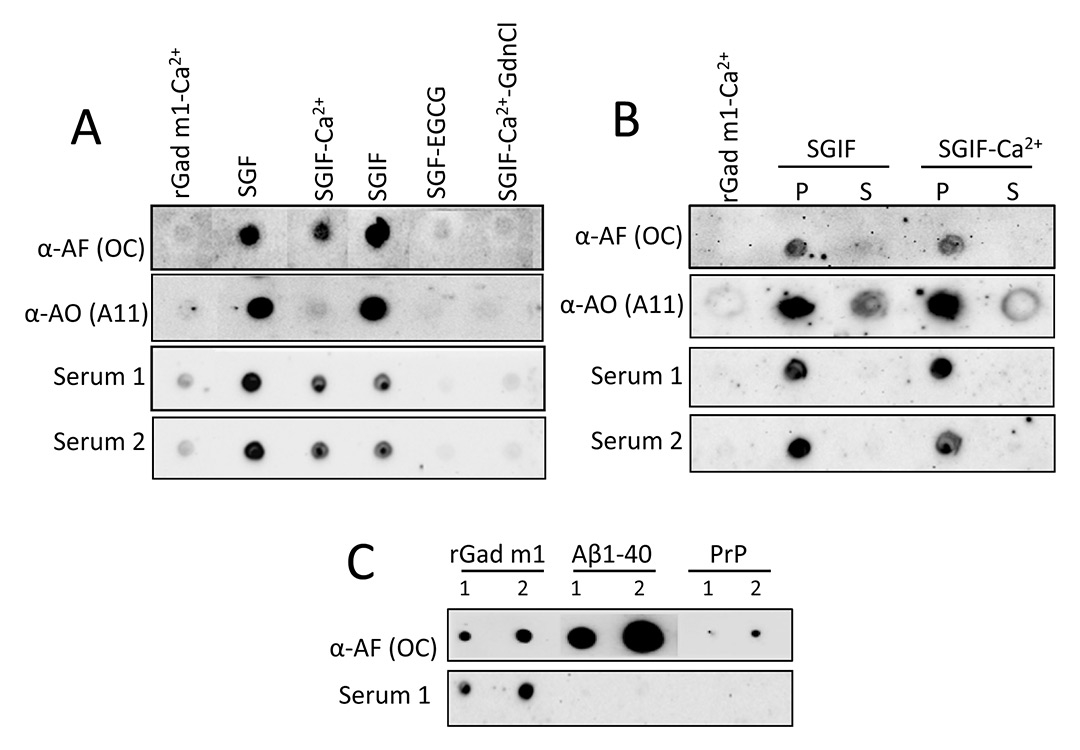
Figure 4
IgE from fish allergic patients sera recognises the rGad m 1 amyloid fibrils. (A) Dot-blot analysis of the immunoreactivity of rGad m 1 species probed with anti-amyloid fibrils (OC), anti-amyloid oligomer (A11) antibodies and with the sera from two fish-allergic patients indicate IgE binding to native amyloid fibrils. Dots correspond to 20 ng of fresh rGad m 1 solution in simulated intestinal fluid (SIF) containing 5 mM Ca2+, rGad m 1 fibrillated for 36 h in simulated gastric fluid (SGF) and then diluted 1/50 in SGF, SIF with 5 mM EDTA (SGIF), SIF with 5 mM CaCl2 (SGIF-Ca2+) and SIF with 5 mM CaCl2 and 6 M GdnCl. SGF- epigallocatechin gallate (EGCG) corresponds to rGad m1 incubated in the presence of 20 μM EGCG for 36 h and then diluted 1/50 in SGF. (B) Separation of soluble (S) and pellet (P) fractions reveal that allergic patients IgE recognise insoluble amyloid fibrils. Fibrils formed at 140 μM protein concentration in SGF were diluted 1/50 in SIF either with 5 mM EDTA (SGIF) or 5 mM CaCl2 (SGIF-Ca2+) and centrifuged at 100,000xg for 1 h at 4 °C. (C) Anti-amyloid fibrils antibody (OC) reacts with amyloid assemblies of rGad m 1, Aβ1–40 and HaPrP 23–231, but IgE from allergic patients sera recognised specifically rGad m 1 fibrils. Labels 1 and 2 correspond to the load of 10 and 20 ng of protein, respectively.
To adopt the amyloid conformation, proteins must contain segments which are able to form a steric zipper [17, 18]. These segments must be exposed to solvent and achieve a local concentration high enough to overcome the entropy of fibre formation [14–16]. We first asked whether β-parvalbumin sequences from fish with commercial value contain adhesive hexapeptides forming steric zippers using the Zipper Db algorithm [17]. All sequences tested displayed at least two regions with adhesive properties, mainly overlapping A, E and F helical regions (fig. 1). Importantly, the adhesive sequence AETKAF located at helix E and linking IgE epitopes is highly conserved and only some herring and mackerel sequences do not have it (fig. 1).
Once the theoretical capacity had been identified, to test amyloid formation we selected Atlantic cod β-parvalbumin (Gad m 1) as a model given its high IgE cross reactivity, and assayed the binding of the amyloid-specific dye thioflavin T (ThT), the formation of SDS-resistant aggregates by means of SDS-PAGE, the presence of β-sheet structures measured with circular dichroism (CD) spectroscopy, and the presence of fibrils in atomic force microscopy (AFM) images. These analyses were performed while simulating the gastric (SGF: 50 mM HCl-Gly pH 2, 35 mM NaCl) and the intestinal (SIF: 50 mM Tris-HCl pH 7.4, 35 mM NaCl) fluids, both in the presence of 5 mM EDTA or 5 mM CaCl2+ to account for cation-free and bound forms. Recombinant Gad m 1 showed fast fibrillation under intestinal conditions in the presence of EDTA as assessed with ThT binding (fig. 2A), solubility assays (fig. 2B), SDS-PAGE (fig. 2C), CD (fig. 2D) and AFM (fig. 2E). At acid pH where Ca2+ binding is impeded by protonation of the side chains involved in its binding site, rGad m 1 aggregated both in the presence of EDTA or Ca2+ (fig. 2A‒E). Aggregation is initiated from the α-helix-poor conformation of the apoform that evolves into a β-sheet-rich conformation (fig. 2D). Under these conditions fibrils were notably present, but aggregation was predominated by lentil shaped oligomers (oblate ellipsoids) about 2 nm high and 40 nm wide. Since transit through the gastrointestinal tract involves exposure first to pH 2 and then to pH >6.5, we simulated the process by performing a 10-min preincubation at pH 2 followed by dilution to pH 7.4. Under these conditions, rGad m 1 formed fibrils in the presence of both EDTA and Ca2+ (fig. 2A‒E). Importantly, exposure to pH <2.5 plays a key role in permitting Ca2+-bound rGad m 1 to fibrillate, since incubation at pH >4.5 impeded the process (fig. 2A, fig. 2E). On the other hand, the slower rate and efficiency observed in the presence of Ca2+ compared with EDTA under simulated gastrointestinal conditions suggest competition between aggregation and cation binding.
To assess whether amyloid formation confers protease resistance, we analysed the rGad m 1 proteolytic pattern under GIT-relevant conditions (fig. 3A). In SGF (pH 2) containing either EDTA or Ca2+ at 5 mM, rGad m 1, freshly dissolved or preincubated for 36 h in such media, resists the action of pepsin and generates full length and truncated chains. Increasing the pH to 7.4 and adding proteinase K instead of the mixture of trypsin and chymotrypsin to minimise the strong Ca2+ dependence of protease activity, showed no further digestion. On the contrary, when rGad m 1 was treated with 20 μM of the anti-amyloid compound epigallocatechin gallate (ECGC) [33] to impair fibril assembly (fig. 3B), acid and neutral digestion proceed with significantly higher efficiency (fig. 3A).
To demonstrate that amyloid fibrils function as depots and release species, we performed a fibre release assay under GIT-relevant conditions [34]. For this purpose, rGad m 1 fibrils were harvested by centrifugation and diluted in SIF, both in the presence and absence of Ca2+, and the process was analysed by means of ThT binding (fig. 3C), CD (fig. 3D) and DLS (fig. 3E). Incubation of fibrils in SIF containing EDTA concurs with about 50% reduction of ThT fluorescence and minor changes in the CD spectrum, indicating the dissociation of fibrils into entities with similar secondary structure. DLS analysis of the supernatant of a 100,000 x g centrifugation revealed, as function of protein concentration, species with RH of 2.2 ± 0.2 nm, 4.5 ± 0.5 nm and 21 ± 0.5 nm which correspond to Ca2+-free monomers and oligomers. In contrast, fibrils incubated in SIF with Ca2+ underwent a larger reduction in ThT binding and changes in the CD indicating an increase in α-helix structure. In this case, the supernatant contained only Ca2+-bound (RH = 1.85 ± 0.1 nm) and Ca2+-free (RH = 2.2 ± 0.2 nm) monomers. Taken together, these results indicated that rGad m 1 fibrils release oligomers and monomers, whose relative populations depend on Ca2+ availability and the protein concentration. Importantly, both amyloid fibrils and oligomers are membrane-active structures with the ability to increase permeability, which could play a role in facilitation of passage of the allergen through the intestinal epithelium, which is required for the sensitisation and the effector phases [35, 36].
To assess the allergenicity of the different rGad m 1 species (monomers, oligomers and fibrils) we tested their IgE recognition by means of dot blot analysis using sera of two patients allergic to fish and sensitised to fish parvalbumin (fig. 4A). Unexpectedly, all forms involving presence of amyloid aggregates were recognised by the sera of fish-allergic patients with an affinity pattern similar to that of the anti-amyloid fibrils OC antibody but different from that of anti-oligomers A11 antibody. Indeed, IgE reactivity was associated with the insoluble aggregates isolated in the pellet fraction of a 100,000 x g centrifugation (fig. 4B) and was prevented by incubation with the anti-amyloid compound EGCG and by their denaturation with GdnCl (fig. 4A). Moreover, IgE reactivity is specific for rGad m 1 amyloids since Aβ1–40 and PrP fibrils displaying anti-amyloid fibril OC antibody reactivity were not recognised by patient sera (fig. 4C). These data indicated that serum recognition involved amyloid assembly and resulted from a fibril-induced protein specific epitope (fig. 4A).
Discussion
Despite the large variety of proteins present in foods, only a few of them are capable of sensitising and eliciting an IgE response [1–3]. These food allergens are mainly present in milk, egg, peanut, wheat, fish and shellfish, and consist of a limited number of protein families with various functions (hydrolases, binding and transport of ligands, storage and cytoskeleton scaffolds) [11, 37, 38]. Food allergens undergo large environmental changes during digestion, triggering profound structural alterations that can be crucial for their pathogenicity. Formation of amyloid aggregates under GIT conditions has been reported for several model proteins, some of which, such as ovalbumin, β-lactoglobulin and lysozyme, behave as food allergens [34, 39, 40–42]. It must be stressed that the amyloid structural signature was established in protein extracts rich in ovalbumin and legumin [39]. Here, we show that rGad m 1, the major type I fish allergen, also forms amyloids in the GIT, and that these assemblies are crucial for allergenicity. This amyloidogenesis explains the role of the GIT, the concentration and ligand dependence, the degradation resistance, the facilitated passage across the intestinal epithelial barrier and the architecture of the IgE epitope.
Aggregation of fish parvalbumin into fibrils occurs through its apo-form, which is consistent with the proamyloid properties of the ligand-free state of proteins such as apomyoglobin and apolipoproteins [43, 44]. It also suggests that the milk allergen apo-Bos d 5 may undergo a similar aggregation event [45]. For fish parvalbumin, the proamyloid form is stabilised by either acid pH or chelates, indicating that gastric pH and food composition (citrates, polyanions, etc) may regulate the efficiency of the aggregation process. On the other hand, the dependence of amyloid formation, both seeding and growing, on the monomer concentration also agrees with the reported variations in pepsin resistance and in the allergenicity of different fish species [28]. Transit through the intestine increases the pH, conditions that favour fibrillation of rGad m 1. At this stage, Ca2+ availability (ligand in general), which depends on its concentration and on the presence of other entities binding it, modulates the efficiency of the dissociation process and the species thus produced.
By virtue of their membrane activity, fibrils and oligomers can cooperate in the disruption of the intestinal barrier permeability, and facilitate the passage of fibrillar oligomers to initiate the sensitisation route. Indeed, these amyloid assemblies share with mucosal adjuvants such as cholera toxin (CT) a deleterious lipid binding activity [6, 35, 36, 46]. On the other hand, as shown for the amyloid-β in retinal pigment epithelium cells, amyloid aggregates could also play a key role in stimulating intestinal epithelial cells to produce IL-33 and trigger type 2 responses, overcoming tolerance and provoking sensitisation [6, 47]. Other amyloid aggregates as Salmonella entericaserovar Typhimurium curli fibrils activate the Toll-like receptor 2 / phosphatidylinositol 3-kinase pathway, enhancing the intestinal epithelial barrier function, which is also tightly regulated by gut microbiota [48–50].
The finding of the amyloid assembly as the Gad m 1 architecture recognised by the IgE sera of patients allergic to fish adds new functions for the amyloid fold and novel routes for fish allergy intervention. Although the amyloid fold was initially recognised in the context of toxic functions, its functional repertoire has been expanded to include storage, coating, catalysis and acquired inheritance roles [12, 14–18]. Fish parvalbumin amyloid adds to the list the function of scaffolding the IgE epitopes of this type I food allergen. The search for the IgE epitopes in fish parvalbumins has yielded several segments around Ca2+-stabilised EF-hands, by use of antigens both denatured proteins and peptides [29–31]. These regions fail to accomplish the multivalence required to cross-link IgE-FcεRI complexes on effector cells in sensitised patients and hence triggering the release of allergy mediators. On the contrary, the structural repetitiveness of the amyloid polymer provides the basis for such multivalence. Generalisation of this finding to other type I food allergens requires the consideration of the insoluble and polymeric protein forms and the use of native conditions during IgE recognition testing. Furthermore, the feasibility of epigallocatechin gallate for preventing rGad m 1 fibrillation suggests a role of anti-amyloid compounds present in food or supplied by the gastrointestinal microbiota as the natural antidotes to food allergens.
Acknowledgements:We thank Drs Manuel Espinosa-Urgel, Rafael Giraldo, Douglas V. Laurents, Victor Muñoz, Rosalía Rodríguez and Silvia Zorrilla for share of reagents and critical reading of the manuscript. JM was supported by a FPI-research contract, MC by a Juan de la Cierva Postdoctoral contract, and AMFE is a Ramón y Cajal fellow.
References
1 Nwaru BI, Hickstein L, Panesar SS, Roberts G, Muraro A, Sheikh A. Prevalence of common food allergies in Europe: a systematic review and meta-analysis. Allergy. 2014;69:992–1007.
2 Muraro A, Werfel T, Hoffmann-Sommergruber K, Roberts G, Beyer K, Bindslev-Jensen C, et al. EAACI food allergy and anaphylaxis guidelines: diagnosis and management of food allergy. Allergy. 2014;69:1008–25.
3 Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133:291–307; quiz 8.
4 Akdis CA. Therapies for allergic inflammation: refining strategies to induce tolerance. Nature Med. 2012;18:736–49.
5 Palomares O. The role of regulatory T cells in IgE-mediated food allergy. J Investig Allergol Clin Immunol. 2013;23:371–82; quiz 2 p preceding 82.
6 Oyoshi MK, Oettgen HC, Chatila TA, Geha RS, Bryce PJ. Food allergy: Insights into etiology, prevention, and treatment provided by murine models. J Allergy Clin Immunol. 2014;133:309–17.
7 Taylor SL, Lehrer SB. Principles and characteristics of food allergens. Crit Rev Food Sci Nutr. 1996;36 Suppl:S91–118.
8 Traidl-Hoffmann C, Jakob T, Behrendt H. Determinants of allergenicity. J Allergy Clin Immunol. 2009;123:558–66.
9 Astwood JD, Leach JN, Fuchs RL. Stability of food allergens to digestion in vitro. Nat Biotechnol. 1996;14:1269–73.
10 Chapman MD, Pomes A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. J Allergy Clin Immunol. 2007;119:414–20.
11 Aalberse RC. Structural biology of allergens. J Allergy Clin Immunol. 2000;106:228–38.
12 Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, et al. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35:349–58.
13 Concha-Marambio L, Diaz-Espinoza R, Soto C. The extent of protease resistance of misfolded prion protein is highly dependent on the salt concentration. J Biol Chem. 2014;289:3073–9.
14 Tycko R, Wickner RB. Molecular structures of amyloid and prion fibrils: consensus versus controversy. Acc Chem Res. 2013;46:1487–96.
15 Greenwald J, Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18:1244–60.
16 Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–203.
17 Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proceed Natl Acad Sci U S A. 2010;107:3487–92.
18 Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–32.
19 Sanchez L, Madurga S, Pukala T, Vilaseca M, López-Iglesias C, Robinson CV, et al. Abeta40 and Abeta42 amyloid fibrils exhibit distinct molecular recycling properties. J Am Chem Soc. 2011;133:6505–8.
20 O’Nuallain B, Wetzel R. Conformational Abs recognizing a generic amyloid fibril epitope. Proc Natl Acad Sci U S A. 2002;99:1485–90.
21 Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18.
22 Martinez J, Lisa S, Sanchez R, Kowalczyk W, Zurita E, Teixidó M, et al. Selenomethionine incorporation into amyloid sequences regulates fibrillogenesis and toxicity. PloS One 2011;6:e27999.
23 Makarava N, Baskakov IV. Purification and fibrillation of full-length recombinant PrP. Methods Mol Biol. 2012;849:33–52.
24 Lisa S, Meli M, Cabello G, Gabizon R, Colombo G, Gasset M. The structural intolerance of the PrP alpha-fold for polar substitution of the helix-3 methionines. Cell Mol Life Sci. 2010;67:2825–38.
25 Lisa S, Domingo B, Martinez J, Gilch S, Llopis JF, Schätzl HM, et al. Failure of prion protein oxidative folding guides the formation of toxic transmembrane forms. J Biol Chem. 2012;287:36693–701.
26 Kuehn A, Swoboda I, Arumugam K, Hilger C, Hentges F. Fish allergens at a glance: variable allergenicity of parvalbumins, the major fish allergens. Front Immunol. 2014;5:179.
27 Moraes AH, Ackerbauer D, Kostadinova M, Bublin M, de Oliveira GA, Ferreira F, et al. Solution and high-pressure NMR studies of the structure, dynamics, and stability of the cross-reactive allergenic cod parvalbumin Gad m 1. Proteins. 2014;82:3032–42.
28 Griesmeier U, Vazquez-Cortes S, Bublin M, Radauer C, Ma Y, Briza P, et al. Expression levels of parvalbumins determine allergenicity of fish species. Allergy. 2010;65:191–8.
29 Elsayed S, Apold J. Immunochemical analysis of cod fish allergen M: locations of the immunoglobulin binding sites as demonstrated by the native and synthetic peptides. Allergy. 1983;38:449–59.
30 Untersmayr E, Szalai K, Riemer AB, Hemmer W, Swoboda I, Hantusch B, et al. Mimotopes identify conformational epitopes on parvalbumin, the major fish allergen. Mol Immunol.2006;43:1454–61.
31 Perez-Gordo M, Pastor-Vargas C, Lin J, Cases B, Ibáñez MD, Vivanco F, et al. Epitope mapping of the major allergen from Atlantic cod in Spanish population reveals different IgE-binding patterns. Mol Nutr Food Res. 2013;57:1283–90.
32 Swoboda I, Balic N, Klug C, Focke M, Weber M, Spitzauer S, et al. A general strategy for the generation of hypoallergenic molecules for the immunotherapy of fish allergy. J Allergy Clin Immunol. 2013;132:979–81 e1.
33 Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, et al. Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington’s disease models. Hum Mol Genet. 2006;15:2743–51.
34 Tufail S, Owais M, Kazmi S, Balyan R, Kaur Khalsa J, Faisal SM, et al. Amyloid form of Ovalbumin evokes native antigen specific immune response in the host: prospective immuno-prophylactic potential. J Biol Chem. 2014.
35 Last NB, Miranker AD. Common mechanism unites membrane poration by amyloid and antimicrobial peptides. Proc Natl Acad Sci U S A. 2013;110:6382–7.
36 Reynolds NP, Soragni A, Rabe M, Verdes D, Liverani E, Handschin S, et al. Mechanism of membrane interaction and disruption by alpha-synuclein. J Am Chem Soc. 2011;133:19366–75.
37 Hoffmann-Sommergruber K, Mills EN. Food allergen protein families and their structural characteristics and application in component-resolved diagnosis: new data from the EuroPrevall project. Anal Bioanal Chem. 2009;395:25–35.
38 Breiteneder H, Mills EN. Molecular properties of food allergens. J Allergy Clin Immunol. 2005;115:14–23.
39 Astbury WT, Dickinson S, Bailey K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J. 1935;29:2351–601.
40 Lara C, Gourdin-Bertin S, Adamcik J, Bolisetty S, Mezzenga R. Self-assembly of ovalbumin into amyloid and non-amyloid fibrils. Biomacromolecules. 2012;13:4213–21.
41 Jones OG, Adamcik J, Handschin S, Bolisetty S, Mezzenga R. Fibrillation of beta-lactoglobulin at low pH in the presence of a complexing anionic polysaccharide. Langmuir. 2010;26:17449–58.
42 Mulaj M, Foley J, Muschol M. Amyloid oligomers and protofibrils, but not filaments, self-replicate from native lysozyme. J Am Chem Soc. 2014;136:8947–56.
43 Fandrich M, Fletcher MA, Dobson CM. Amyloid fibrils from muscle myoglobin. Nature. 2001;410:165–6.
44 Hatters DM, Howlett GJ. The structural basis for amyloid formation by plasma apolipoproteins: a review. Eur Biophys J. 2002;31:2–8.
45 Roth-Walter F, Pacios LF, Gomez-Casado C, Hofstetter G, Roth GA, Singer J, et al. The major cow milk allergen Bos d 5 manipulates T-helper cells depending on its load with siderophore-bound iron. PloS One 2014;9:e104803.
46 Lencer WI, Tsai B. The intracellular voyage of cholera toxin: going retro. Trends Biochem Sci. 2003;28:639–45.
47 Liu XC, Liu XF, Jian CX, Li CJ, He SZ. IL-33 is induced by amyloid-beta stimulation and regulates inflammatory cytokine production in retinal pigment epithelium cells. Inflammation. 2012;35:776–84.
48 Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–12.
49 Oppong GO, Rapsinski GJ, Newman TN, Nishimori JH, Biesecker SG, Tukel C. Epithelial cells augment barrier function via activation of the Toll-like receptor 2/phosphatidylinositol 3–kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infect Immun. 2013;81:478–86.
50 Cao S, Feehley TJ, Nagler CR. The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS letters 2014;588:4258–66.



