The Lausanne Institutional Biobank: A new resource to catalyse research in personalised medicine and pharmaceutical sciences
DOI: https://doi.org/10.4414/smw.2014.14033
Vincent
Mooser, Christine
Currat
Summary
Breakthrough technologies which now enable the sequencing of individual genomes will irreversibly modify the way diseases are diagnosed, predicted, prevented and treated. For these technologies to reach their full potential requires, upstream, access to high-quality biomedical data and samples from large number of properly informed and consenting individuals and, downstream, the possibility to transform the emerging knowledge into a clinical utility.
The Lausanne Institutional Biobank was designed as an integrated, highly versatile infrastructure to harness the power of these emerging technologies and catalyse the discovery and development of innovative therapeutics and biomarkers, and advance the field of personalised medicine. Described here are its rationale, design and governance, as well as parallel initiatives which have been launched locally to address the societal, ethical and technological issues associated with this new bio-resource.
Since January 2013, inpatients admitted at Lausanne CHUV University Hospital have been systematically invited to provide a general consent for the use of their biomedical data and samples for research, to complete a standardised questionnaire, to donate a 10–ml sample of blood for future DNA extraction and to be re-contacted for future clinical trials. Over the first 18 months of operation, 14,459 patients were contacted, and 11,051 accepted to participate in the study.
This initial 18–month experience illustrates that a systematic hospital-based biobank is feasible; it shows a strong engagement in research from the patient population in this University Hospital setting, and the need for a broad, integrated approach for the future of medicine to reach its full potential.
The potential of genomics to transform medicine
Sequencing the 3–billion base-pairs of an individual genome for less than 1000 CHF has now become reality. The first human genome was published in 2003. Four years later, a seminal study utilising a technology to analyse and interrogate a couple of million of common single nucleotide polymorphisms (SNPs) spread over the entire genome (i.e. genome-wide association studies, GWAS [1]) was published. This article was soon followed by hundreds of such publications (http://www.genome.gov/gwastudies/index.cfm?pageid=26525384#searchForm) which have revealed a large number of new chromosomal regions associated with a broad range of medical conditions. Even more recently, new sequencing technologies have unravelled an abundance of rare, functional variants in the human genome, secondary to a recent explosive population growth [2–4]. In a particular series of experiments, sequencing the coding portion of 202 drug target genes in 14,002 individuals (including 3000 individuals from the Lausanne area) revealed more than 11,000 amino-acid changing variants, with the vast majority of them (74%) being novel and present in only one or two individuals; moreover almost half of these rare variants were predicted to be functional with damaging effects on the gene product. In that sense, these rare variants generate “human knock-out/down” effects if they lead to loss or reduction of function of their gene product or if they have a dominant negative effect; conversely, they may lead to “human over-expressors” of the gene, in case of gain-of-function mutations or excessive gene transcription, for instance in the presence of large number of gene copies [5].
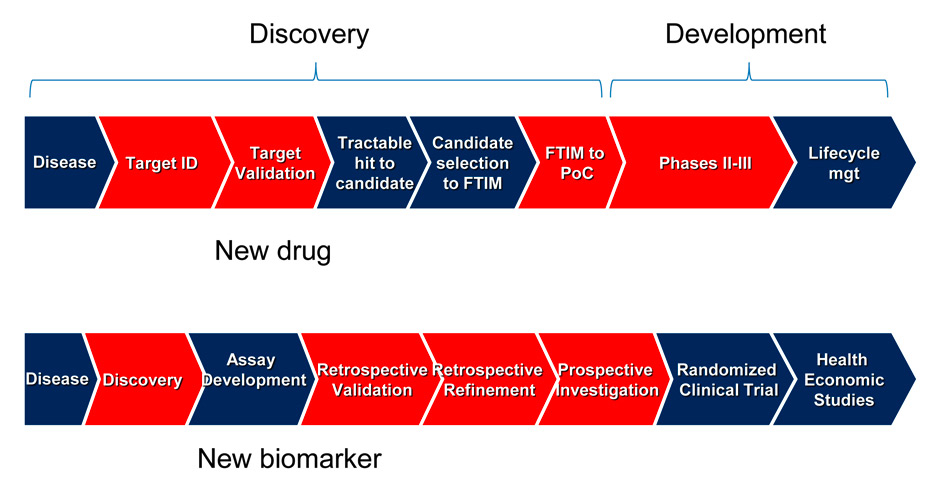
Figure 1
Chevron diagrams showing the various steps which lead to the discovery and development of new drugs (upper panel) and biomarkers (lower panel).
Steps where genomic information and large bio-resources can be used to help decision making and accelerate the process of discovery and development are shown in red. Target ID: target identification; FTIM = first time into man; PoC = proof-of-concept. Upper part of the chevron diagram adapted from [9].
These rare “experiments by nature” represent a blessing for biomedical research [6]. They open up unprecedented opportunities to accelerate the discovery and the development of innovative therapeutics and biomarkers and, at term, to tailor medical interventions to the particular needs and the specific genetic make-up of individuals [7]. This potential is further accelerated when genetics is combined with electronic medical records or other sources of phenotypic data [8]. Bringing new drugs to the market obeys to a series of well-defined steps [9], and shares many features in common with the discovery and development of innovative biomarkers [10] (fig. 1). The transition from one step to the next step depends on usually difficult-to-make go/no-go decisions, which are based on information which needs to be gathered using a variety of approaches.
Human genetic information has already proved to be very instrumental in the discovery of new drug targets. Examples to illustrate this point are the observations that humans genetically deficient in CCR5 receptors are resistant to HIV infection, or that individuals lacking PCSK9 have low LDL-cholesterol levels and are protected from heart disease [11, 12]. These genetic observations on “human knock-out” have prompted the discovery of CCR5 inhibitors and PCSK9 inhibitors [13], respectively. Further down the pipeline, statins (which up-regulate the LDL-receptor at the cell surface to accelerate the clearance of LDL particles) were originally tested and were shown to be effective (i.e. in proof of concept studies) on a small number of individuals who were genetically defective in half of their LDL receptors (i.e. heterozygous familial hypercholesterolemia). The commoditisation of large-scale exome and, soon, whole genome sequencing to detect carriers of rare functional mutations opens up the possibility to apply this paradigm to essentially any new therapeutic in development for chronic conditions, with the expectation to lead to shorter, faster and more decisive proof-of-concept studies. However, the problem is that given the fact that these mutations are rare, access to genomic data from large, or very large number of individuals is required.
As is the case for new drugs, the discovery and development of innovative biomarkers becomes a long, expensive, bumpy and risky road going from the discovery of the biomarker to its validation and the demonstration that the marker brings clinical benefit (lower panel fig. 1) [10]. Finally, the prospect of personalising a therapy according to the genetic (or other biomarker) make-up of an individual (i.e. “the right drug at the right dose to the right patient”) is undoubtedly attractive from a conceptual point of view, and has mostly permeated the oncology field. However, major efforts are to be made for personalised medicine to reach maturity and fulfil the expectations of the patients, society and industry [6].
In any case, there is no doubt that, once commoditised, high-throughput technologies capable of analysing the entire genome (i.e. genomics), the metabolic profile (metabolomics), and gene expression (transcriptomics) of a particular liquid or tissue sample, or the environment (exposomics), among other *omic approaches integrated into a “systems medicine” network [14], will irreversibly modify the way diseases are diagnosed, predicted, prevented and treated.
The essential role of population engagement and large bioresources
For the full potential of these *omic technologies to be realised requires that they are “fed”, upstream, with high-quality data and samples from large number of properly informed, engaged, well-phenotyped and consenting individuals (fig. 2). The roles of these technologies are to generate new biological data (like whole genome sequencing) and to store, manipulate and analyse these massive amounts of clinical and biological data (i.e. “Big Data”) to create new medical knowledge. New knowledge doesn’t necessarily mean clinical benefit, however. Robust evidence needs to be constructed to demonstrate the clinical utility of this new knowledge (i.e. clinical benefits for the patients, and for the society). To get there requires downstream of these technologies, the engagement of the population into clinical research and the willingness of the citizens to participate in clinical trials, which remain the necessary stepping stone to demonstrate their clinical utility, be it for new interventions or new biomarkers. Finally, keeping in mind that no new drug or biomarker has reached the market (and the patients) without the engagement of private sectors, innovative public-private partnerships will need to be created to deliver novel ways to diagnose and treat diseases, and to monitor the effect of these interventions.
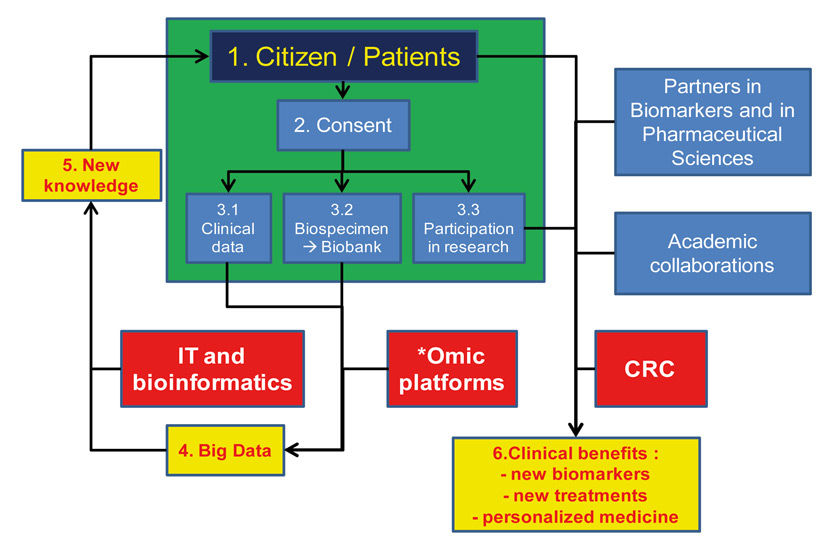
Figure 2
Role of bio-resources like BIL in catalysing translational research.
Emerging *omic technologies (genomics, transcriptomics, metabolomics and others) require access to high-quality data and samples from large number of individuals engaged in research and properly consented for the use of their data and specimen. These technologies generate large amounts of data (i.e. Big Data), which are converted into new knowledge thanks to information technology (IT) and bio-informatics. For their clinical utility to be demonstrated, evidence needs to be built, using clinical research on engaged citizens and in partnership with other academic groups within the private sector. The perimeter of the BIL is illustrated using the green rectangle. CRC = clinical research centre.
The Lausanne Institutional Biobank (Biobanque Institutionnelle de Lausanne, BIL) was designed to harness the power of these emerging *omic technologies, and to facilitate selected steps in the discovery and development of new drugs and biomarkers and in the construction of personalised medicine. More specifically, a major ambition of the BIL is to “feed”, upstream, these technologies with high-quality data and samples, and transform, downstream, the new medical knowledge into benefits for the patients and the society (fig. 2).
The Lausanne Institutional Biobank (BIL): design and governance
The construction of large biobanks like the UK Biobank [15], or smaller bio-resources like CoLaus [16] or the BIL (http://www.chuv.ch/biobanque) raises a series of issues which pertain, among others, to the recruitment of the patients, the access to high-quality medical and phenotypic data, the legal and ethical framework, the protection of private data, as well as funding and economical aspects (fig. 3). The BIL investigators had already participated in the design, implementation and exploitation of the CoLaus Study and the Lausanne Oncology Biobank, and had already experienced that the local population was engaged and interested in participating in biomedical research.
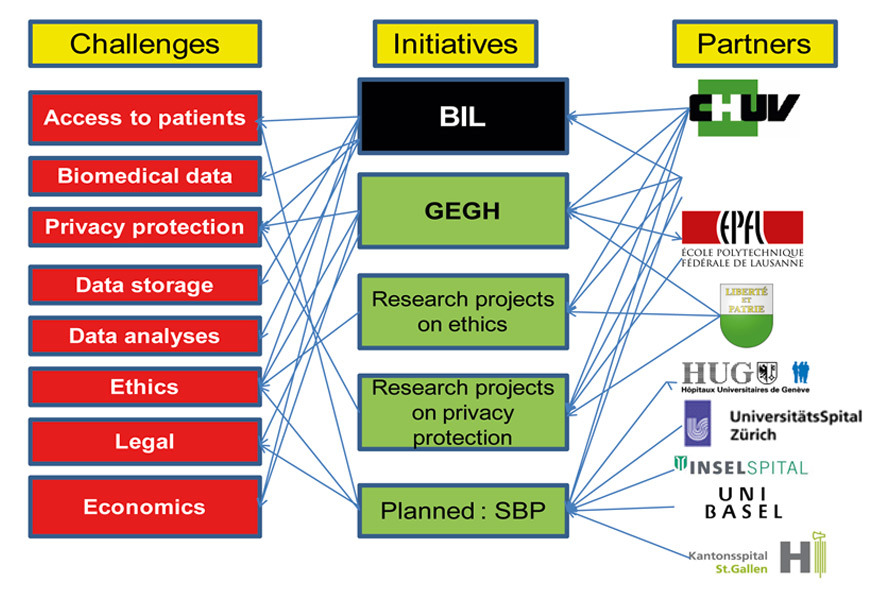
Figure 3
Landscape of the Lausanne Institutional Biobank (BIL).
Selected challenges associated with the construction of a bio-resource like the BIL are listed on the left-hand side. Partners in addressing these challenges are listed on the right-hand side and the BIL and parallel projects are listed in the middle. GEGH = Groupe d’Experts sur le Génome Humain (http://www.vd.ch/actualite/articles/experts-en-medecine-genomique/). SBP = Swiss Biobanking Platform, at the planning stage. An overview of the local activities on the societal aspects of genomic medicine, ethics, privacy protection and personalised medicine is provided in references [17–19] and [7].
The BIL project was designed following extensive discussions with health care professionals and experts in biomedical sciences, ethicists and lawyers (more particularly within the frame of the new Law on Research in Humans and its Ordinances, which were enacted in January 1st, 2014), and after benchmarking with similar initiatives in the USA, UK and Asia. It received approval from the Ethics Committee of the Vaud Canton in October 2012. Upon launching of the BIL project, three parallel initiatives were started in the Lausanne area, sponsored by the local government and the UNIL University (i.e. the creation of an Expert Group in Genome Sciences (GEGH, http://www.vd.ch/actualite/articles/experts-en-medecine-genomique/)) [17] and research projects on the ethical aspects of the BIL General Consent [18], and on privacy protection [19]. In parallel, a project is under construction for a Swiss National Biobanking Platform (SBP), under the auspices of the Swiss National Science Foundation, at the interface with the five University Hospitals in Switzerland, the ETH domain and the other Swiss Universities. The initial phase of the BIL project, which has been identified as a key strategic priority by the CHUV Hospital (http://www.chuv.ch/chuv_planstrat201418.pdf), the Lausanne University and the Vaud Ministry of Health and Social Welfare, has been funded jointly by these former two institutions for 2014–2015. Upon careful evaluation, it has recently been decided that the Biobank shall be integrated with the Clinical Research Centre and a dedicated IT group, into the newly created and funded Platform to Support Clinical Research at CHUV/UNIL.
The design of the BIL is conceptually simple. Each patient admitted for hospitalisation in the CHUV University Hospital, is informed about the project and is invited to sign the General Consent for research, to complete a standardised questionnaire and to donate a 10–ml blood sample for future DNA extraction, and consent for future contacts.
The General Consent (http://www.chuv.ch/biobanque/bil_home/bil-patients-famille/bil-participer/bil-le_consentement_general.htm) was developed to facilitate the engagement of participants in clinical research. Individuals consent for the broad use of their biomedical data for research, without having to be re-contacted and re-consented for new projects, as long as these projects have been approved by the local Ethics Committee. In addition, they consent for their genome to be fully sequenced in the future, and accept or not to be re-contacted in case clinically actionable mutations were to be found once their genome had been sequenced, and for future clinical research. They are invited to complete a questionnaire, with standard questions related to their personal history, their family history, and phenotypic variables. Given the new Law on Research on Humans, each patient needs to be informed personally and to provide written consent (i.e. “opt-in” recruitment). A team of dedicated nurses, medical assistants and research personnel has been specifically hired and trained for these tasks. A 10–ml blood sample is collected, usually on the second day of admission, in fasting conditions, in the wards. The blood is transported to the lab where it is spun, generally within 1 hour upon collection. Plasma is isolated and aliquoted into 3 0.5 ml tubes, which are stored at –80 °C. Similarly, the buffy coat is isolated and split into two aliquots, stored in two separate –80 °C freezers for future DNA extraction. A specific biobanking software has been developed for the annotation of the patient’s willingness to participate in research and recording and monitoring of samples type and availabilities.
The strategic direction of the BIL, and the newly created Platform it is part of, is supervised by a Steering Group chaired by the Dean of the Biology and Medical School, and senior representatives from UNIL and CHUV Direction and Department Heads. This group oversees the activity of the BIL operating board. It is also in charge of prioritising which studies will be used as pilots to demonstrate the use of the platform (see below), and to ensure that funding is properly used. A separate board, made of patient’s representatives, is under construction.
Recruitment was launched on January 7th, 2013. Within the first eighteen months (i.e. up to June 30th, 2014), a total of 14,459 patients were contacted and invited to participate in the project, with 11,051 patients consenting to be part of the BIL (i.e. a participation rate of 76%). The basic characteristics of these participants are described in table 1. Experience so far indicates that the approach, which was designed to have the General Consent presented to all inpatients by an independent team of recruiters, provides a significant benefit to the patients, who are better informed about what clinical research means, it facilitates the work of the investigators and diminishes the risk of conflict of interest that investigators may have. The ambition of the BIL is to continue the recruitment at a rate of 10,000 patients per year.
|
Table 1:Characteristics of the first 11,051 participants of the Lausanne Institutional Biobank. |
| n |
11,051 |
| Males (%) / females (%) |
54 /46 |
| Age (mean ± SD) (years) |
60 ± 19 |
| Origin (% Swiss) |
70 |
| Professional status |
40% retired / 26% employee |
| Religion |
37% Catholic / 35% Protestant / 28% other |
|
Table 2: Potential for projects to be performed using the BIL bio-resource. |
|
Type of projects
|
Examples
|
| Characteristics of medical care and practice – Hospital epidemiology |
Impact of drug adverse effects on hospital admissions
Medical care of specific conditions |
| Discovery of genetic and molecular bases of diseases, biomedical traits or response to interventions (including pharmacogenetics) – discovery of novel biomarkers using untargeted *omic technologies |
GWAS analysis of clinical conditions and phenotypic traits [20]
In silico GWAS analyses [8]
Genetic analyses of new phenotypes (e.g. facial morphology [21] |
| Validation of genetic or other biomarkers |
Confirmation of association between genetic and non-genetic markers on biobanked samples from well-characterised individuals |
| Detailed phenotypic analyses on human carriers of null mutations (including PheWAS) (supporting target validation) |
Hormone sensitive lipase [22]
ApoCIII [23]
PCSK9 [11] |
| Proof-of-concept studies for investigational therapeutics tested on carriers of selected genetic mutations or carriers of extreme phenotypes |
Statins in heterozygous familial hypercholesterolemia [24] |
| Genetically enriched Phase IIb/III studies on selected participants of biobanks |
Modelisation in [25] |
| Predictive and preventive medicine |
Breast and ovary cancer prevention in carriers of BRCA1/2 mutations [26] |
The Lausanne Institutional Biobank (BIL): perspectives and opportunities for research
Along the same strategy which underlined the creation of the Swiss HIV Cohort Study, CoLaus (http://www.colaus.ch/en/cls_home/cls_pro_home/cls_pro_publications.htm) or other biobanks, the BIL is constructed to facilitate translational and clinical research for a variety of investigators locally and regionally. Different categories of research projects are envisioned with the BIL, spanning from hospital epidemiology to clinical trials (table 2). These project pertain to the use of biological samples for independent validation of biomarkers, for detailed *omic analyses of selected samples and for enrolment of specific patients into future clinical trials. In parallel, activities have been envisioned with the other Faculties at the Lausanne University, for a global approach to new medicine, with partners at the local (EPFL), regional (University of Geneva) and national level (Swiss Biobanking Platform), for the optimal exploitation of the BIL bio-resource. In addition, creative partnerships are envisioned between this publicly funded initiative and private partners, as a way for “genomic revolution” to benefit the patients and the population.
Acknowledgements:The authors thank the participants of the BIL and the personnel of the BIL: Annick Antille, Anne-Lise Bastian, Laurence Chapatte Delapierre, Pasqualina Corthésy, Réjane Dietrich, Ludovic Dey, Elise Dubois Couture, Annick Ducraux Savoy, Didier Foretay, Laurence Gander, Vanessa Jaquet, Amédée Kibalabala, Naomi Kramer, Chiyama Mathivathanasekaram, Carole Morel, Dominique Niksch, Anne-Laure Nicoulaz, Julia Parafita, Karine Pierre, Maria Ramos Varela, Claudia Rochat, Cindy Roth, Audrey Roth and Catherine Saner Zilian. Finally, they express their gratitude to the General Management of the CHUV Hospital, the Rectorat of the Lausanne University (UNIL) and the Décanat of the Biology and Medical School (FBM) for their continuous support and for the initial funding of the recruitment of the BIL. In addition, they thank the members of the Scientific Advisory Board for their engagement and their input: Dr David Baud, M. Urs Benz, Pr Murielle Bochud, Dr Pierre-Yves Bochud , Dr Luisa Bonafé, Dr Mirela Caci, Pr Chin-Bin Eap, M. Darcy Christen, Pr George Coukos, Pr Jean-François Démonet, Pr Nicolas Demartines, Pr Alban Denys , M. Patrick Genoud, M. Stefan Kohler, Pr Lucas Liaudet, Pr Olivier Michielin, M. Alain Petter, Mrs Jeanne-Pascale Simon, Pr Christian Simon, Pr Alexander So, M. Nicolas Rosat, Pr Andrea Superti Furga, Pr Gérard Waeber and Dr Christian Wider.
References
1 Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Human Genet. 2012;90(1):7–24.
2 Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–9.
3 Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, et al. An Abundance of Rare Functional Variants in 202 Drug Target Genes Sequenced in 14,002 People. Science. 2012.
4 Coventry A, Bull-Otterson LM, Liu X, Clark AG, Maxwell TJ, Crosby J, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1(131.
5 Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, et al. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet Part A. 2012;158A(7):1523–5.
6 Ginsburg G. Medical genomics: Gather and use genetic data in health care. Nature. 2014;508(7497):451–3.
7 Mooser V. Genomics and personalized medicine. Praxis. 2014;103(10):567–71.
8 Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–10.
9 Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6(1):29–40.
10 Ziegler A, Koch A, Krockenberger K, Grosshennig A. Personalized medicine using DNA biomarkers: a review. Hum Genet. 2012;131(10):1627–38.
11 Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–5.
12 Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–83.
13 Sullivan D, Olsson AG, Scott R, Kim JB, Xue A, Gebski V, et al. Effect of a Monoclonal Antibody to PCSK9 on Low-Density Lipoprotein Cholesterol Levels in Statin-Intolerant Patients: The GAUSS Randomized Trial. JAMA: the journal of the American Medical Association. 2012:1–10.
14 Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. New biotechnology. 2012;29(6):613–24.
15 Elliott P, Peakman TC, Biobank UK. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int J Epidemiol. 2008;37(2):234–44.
16 Firmann M, Mayor V, Vidal PM, Bochud M, Pecoud A, Hayoz D, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6.
17 Maillard PY. Sequencing the human genome and society. Praxis. 2014;103(10):551–3.
18 Barazzetti G, Kaufmann A, and Benaroyo L. Ethical and social issues associated with genomic medicine. Praxis. 2014;103(10):573–7.
19 Raisaro JL, Ayday E, Hubaux JP. Patient privacy in the genomic era. Praxis. 2014;103(10):579–86.
20 Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–13.
21 Windhager S, Schaschl H, Schaefer K, Mitteroecker P, Huber S, Wallner B, Fieder M. Variation at genes influencing facial morphology are not associated with developmental imprecision in human faces. PLoS One. 2014;9(6):e99009.
22 The TG, Hdl Working Group of the Exome Sequencing Project NHL, Blood I. Loss-of-Function Mutations in APOC3, Triglycerides, and Coronary Disease. N Engl J Med. 2014;371(1):22–31.
23 Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Loss-of-Function Mutations in APOC3 and Risk of Ischemic Vascular Disease. N Engl J Med. 2014;371(1):32–41.
24 Mabuchi H, Haba T, Tatami R, Miyamoto S, Sakai Y, Wakasugi T, et al. Effect of an inhibitor of 3–hydroxy-3–methyglutaryl coenzyme A reductase on serum lipoproteins and ubiquinone-10–levels in patients with familial hypercholesterolemia. N Engl J Med. 1981;305(9):478–82.
25 Hu Y, Li L, Ehm MG, Bing N, Song K, Nelson MR, Talmud PJ, et al. The benefits of using genetic information to design prevention trials. Am J Hum Genet. 2013;92(4):547–57.
26 Couch FJ, Nathanson KL, Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014;343(6178):1466–70.


