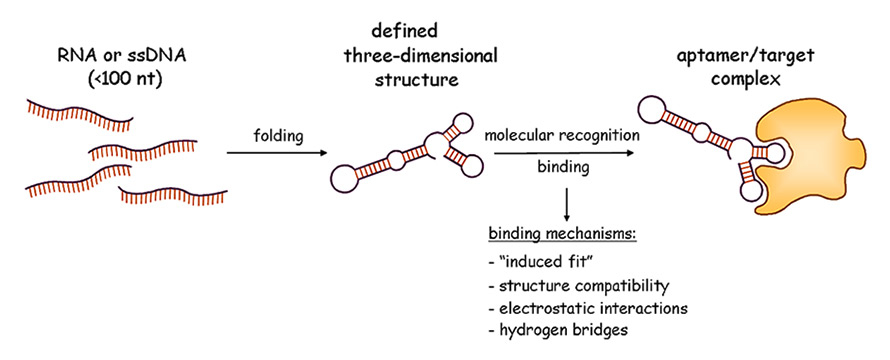
Figure 1
Schematic view of the aptamer molecular recognition principle (Graphic: Regina Stoltenburg, UFZ).
nt = nucleotide; RNA = ribonucleic acid; ssDNA = single-stranded deoxyribonucleic acid
DOI: https://doi.org/10.4414/smw.2014.13908
Abbreviations
AS1411 (AGRO100) experimental anticancer DNA aptamer
CCRF-CEM acute lymphoblastic T-cell leukaemia cell line
CT computed tomography
DNA deoxyribonucleic acid
dsDNA double-stranded deoxyribonucleic acid
FRET fluorescence resonance energy transfer
G guanine
GI-1 human glial cell line
GRO G-rich oligonucleotides
LNA locked nucleic acid
MUC1 DNA aptamer for epithelial tumour marker mucin 1
PCR polymerase chain reaction
PEG polyethylene glycol
PLGA poly(D,L-lactic-co-glycolic acid)
PSMA prostate-specific membrane antigen
RNA ribonucleic acid
RT-PCR reverse transcription polymerase chain reaction
SELEX systematic evolution of ligands by exponential enrichment
SK-BR-3, MCF-7 breast cancer cell lines
ssDNA single-stranded deoxyribonucleic acid
Aptamers are single-stranded deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) oligonucleotides, which are able to bind their target with high selectivity and affinity. Owing to their multiple talents, aptamers are nanosystems well qualified for the development of new biomedical devices for analytical, imaging, drug delivery and many other medical applications. For instance, the common use of nanoparticles for drug delivery and bioimaging in cancer diagnostics and treatment can be substantially improved by modification with aptamers to enhance the specific binding of the nanoparticles via the specific aptamer binding to the target molecule. Because of their target affinity, aptamers can direct the transport of the aptamer-nanoparticle conjugate to this target. The subsequent aptamer binding to the target “anchors” the nanoparticle-aptamer conjugate at its site of action. Furthermore, aptamer-nanoparticle conjugates can improve the measurement of cancer-relevant parameters and, hence, contribute to the development of intelligent medical devices. Despite their advantages and unique properties as molecular probes, however, aptamers have been used sparingly for medical applications because of the limited number of aptamers available for medically relevant target molecules.
This article gives a brief overview of recent relevant research into aptamers and trends in their use in cancer medicine. A description of aptamers, their development and functionalities related to nanoparticle modification is given. The main part of the article is dedicated to the current developments of aptamer-modified nanoparticles and their use in cancer research, diagnostics and treatment.
Aptamers are artificial nucleic acid ligands. They are short single-stranded DNA (ssDNA) or RNA oligonucleotides that bind to their targets with high selectivity and sensitivity because of their three-dimensional shape. The word “aptamer” was invented by Andrew Ellington and coworkers. It derived from the Latin “aptus” (fitting) and meant a nucleotide polymer that fits to its target [1]. Target molecules for aptamers can be virtually any class of substrate ranging from whole cells to large molecules, like proteins, to peptides to drugs and organic small molecules or even metal ions. Aptamers have the potential for use as specific binding molecules with high affinities in medical and pharmaceutical basic research, drug development, diagnosis and therapy. They can be applied in analytical and separation tools as molecular recognition and binding elements and can be used for the investigation of binding phenomena in proteomics. The technology to evolve aptamers was discovered in 1990 [2, 3] and was named ‘SELEX’ (Systematic Evolution of Ligands by EXponential enrichment) by Larry Gold. SELEXTM is a trademark of Gilead Sciences, Inc., who is the owner of the process patent.

Figure 1
Schematic view of the aptamer molecular recognition principle (Graphic: Regina Stoltenburg, UFZ).
nt = nucleotide; RNA = ribonucleic acid; ssDNA = single-stranded deoxyribonucleic acid
The functionality of aptamers is based on their stable three-dimensional structure, which depends on the primary sequence, the length of the nucleic acid molecule (typically smaller than 100 nucleotides) and the environmental conditions. The specific and complex three-dimensional structures of aptamers are characterised by stems, internal loops, bulges, hairpins, tetra loops, pseudoknots, triplicates, kissing complexes or G-quadruplex structures. Binding of the aptamer to its target (fig. 1) results from a combination of complementarity in the geometrical shape, stacking interactions of aromatic rings and the nucleobases of the aptamers, electrostatic interactions between charged groups, van der Waals interactions and hydrogen bonds [4]. Aptamers are considered to be “nucleic acid antibodies”, sometimes with additional superior properties [5]. Many of the selected aptamers show affinities comparable to those observed for monoclonal antibodies. Aptamers can distinguish between chiral molecules, they are capable of recognising a distinct epitope of a target molecule and differentiate closely related targets (e.g. theophylline and caffeine [6]).
Since the early phase of the SELEX technology, it has often been modified to make the selection process more efficient and less time consuming, or to select aptamers with particular binding features for different target molecules and for different applications [7]. Numerous variants of the original SELEX process have been described. In principle, the SELEX process is characterised by iterative cycles of in-vitro selection and enzymatic amplification. Thereby, the operation mimics a Darwinian type process, driving the selection towards relatively few (but optimised) structural motifs, which show the highest specificities and affinities to the selection target. The starting point of a typical SELEX process is a chemically synthesised random DNA oligonucleotide library which consists of about 1013–1015 different sequence motifs [8]. The DNA library has to be converted into an RNA library prior to the start of the RNA SELEX process. In the first selection round, the complex RNA or DNA pool is incubated with the target. Subsequent stringent washing steps separate unbound and weakly bound oligonucleotides from the resulting bound complexes. Target bound oligonucleotides are eluted and amplified by polymerase chain reaction (PCR) for DNA SELEX or reverse transcription PCR (RT-PCR) for RNA SELEX. The resulting double-stranded DNA (dsDNA) has to be transformed into a new oligonucleotide pool by purifying the relevant ssDNA or by in-vitro transcription and purification of the synthesised RNA. This new enriched pool of selected oligonucleotides is used in the following SELEX round for a binding reaction with the target. Enrichment up to a saturation concentration of target-specific oligonucleotides indicates that the SELEX process is finished. The selected aptamer pool is cloned to obtain individual aptamers and the individual aptamers are sequenced. Sequences are analysed, and representative aptamers are chosen and used in binding assays to characterise their binding features, including the affinities and specificities, in more detail.
Cell-specific aptamers binding to entire cells or to a specific epitope on the cell surface are selected by using whole cells as targets in the SELEX process [9]. Moreover, it is possible to generate aptamers that can differentiate between two types of cells, for example, cancer cells from normal cells [10]. More detailed description of the SELEX process and of the different methodologies is given in [7].
Aptamers interact directly with their targets by a binding reaction. The bound aptamer has an impact on the function of the target molecule. For example, it can inhibit the binding of this molecule to the cognate receptor.
The first aptamer-based drug (Macugen®, for treatment of age-related wet macular degeneration) is based on the PEGylated form of an antivascular endothelial growth factor aptamer [11]. Some more aptamer-based pharmaceuticals are in various stages of development, from preclinical studies to clinical trials [12].
Aptamers will not replace antibodies, particularly in well-established applications. However, they are going to find their own niche of applications [13]. In comparison with antibodies, they have some advantages because of their molecular nature:
– they have high stability (especially DNA aptamers), allowing storage at room temperature;
– they can be heated up to 95°C or exposed to various solvents or harsh environments, and will return to their original confirmation, providing a long shelf-life [14], and low immunogenicity and toxicity;
– chemical modifications are possible to extend their lifetime in biological fluids, to immobilise them on surfaces, to endow them with markers [6, 15], and to “tune” their half-lives to match the indication [13].
To enhance biostability of aptamers, it is possible to introduce chemical modifications. An appropriate method is to modify the ribose components of RNA aptamers, for example by using 2’-O-methyl substituted nucleotides. In this way, RNA is protected from degradation by nucleases [16, 17]. A 3’-capping with streptavidin-biotin, inverted thymidine or several 5’-caps (amine, phosphate, polyethylene glycol (PEG), cholesterol, fatty acids, proteins, etc.) defends oligonucleotides against exonucleases [18, 19]. Locked nucleic acids (LNAs) [20] show great promise in stabilising aptamers because of their substantially increased helical thermostability and excellent mismatch discrimination when hybridised with RNA or DNA. Furthermore, they are resistant to degradation by nucleases. In LNA nucleotides, post-SELEX modified, the sugar is made bicyclic by covalently bridging the 2’-oxygen and the 4’-carbon with methylene [21].
Aptamer-based molecular imaging agents have short circulating half-lives in the body, whereas antibody-based imaging agents may circulate for days to weeks [13]. Antidotes can be rationally designed to reverse the effect of the aptamer molecules. The antidote can bind to the aptamer, disrupt its structure and cause its inactivation. In this way, more control and better timing of reversal of aptamer activity in the body is possible [15].
Sequence truncations are performed to narrow down the binding regions of an aptamer. Regions that are not important for direct interaction with the target are removed. Sometimes such truncations even result in raised affinity [22, 23]. The smaller size of aptamers might enable access to epitopes that are unavailable to antibodies [15].
Since aptamers can be selected for specific binding to small molecules, they are applicable for detecting small molecule ligands, such as cellular metabolites, or can be used for therapeutic drug monitoring.
Because of their outstanding properties, aptamers are used in combination with nanoparticles for biomedical sensing and detection, and as aptamer-nanoparticle conjugates for smart drug delivery. The aptamer-nanoparticle conjugates enable active controlled delivery of drugs that are incorporated in the nanoparticles once they are bound to a disease site because of the aptamer affinity to this site [5].
This research area has already seen substantial growth in recent years [24]. We will exemplify here results in the development of aptamer-based nanomaterials for cancer detection and imaging. In this special field, the progress has been remarkable.
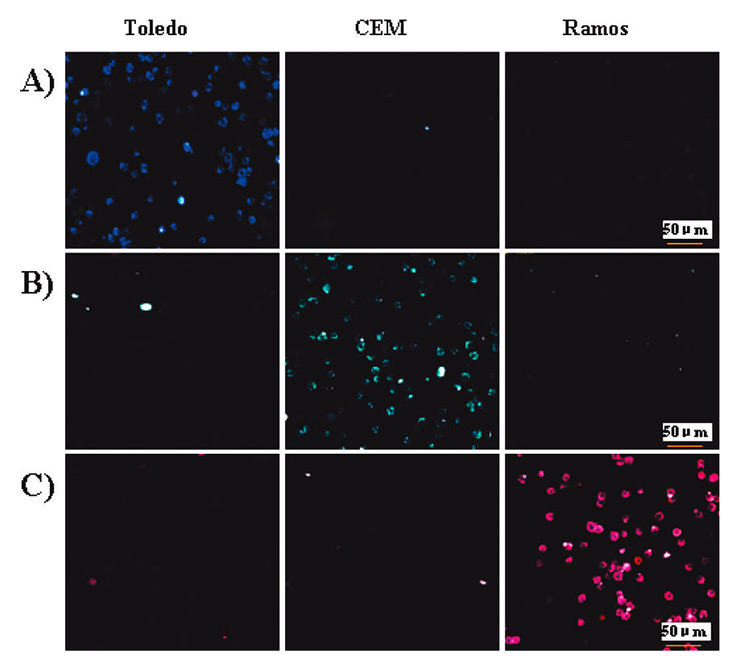
Figure 2
Confocal microscopy images of individual nanoparticle-aptamer conjugates with the three different cells (Toledo, CCRF-CEM, and Ramos): (A) nanoparticle-(FAM)-aptamer T1, specific for Toledo, (B) nanoparticle-(FAM-R6G)-aptamer sgc8, specific for CCRF-CEM, and (C) nanoparticle-(FAM-R6GROX)-aptamer TDO5, specific for Toledo. Reprinted with permission from [35]. Copyright 2009 American Chemical Society.
For exact selection of the proper treatment for cancer, shorter detection and diagnosis timelines are essential. Therefore, the development of quick and easy detection methods to determine the different cancer types is required. Much more than for other diseases, in cancer the consistent multitude of different cancer cells need the implementation of personalised treatment. Molecular recognition of disease-specific biomarkers, especially the recognition of proteins or other biological molecules that differentiate between normal and abnormal cells is a fundamental challenge in cancer cell biology [25]. Aptamers are very suitable for this purpose. Systematic in-vitro development provides aptamers that are able to detect even small differences between molecules and between various cell types. Aptamers have a low molecular weight, fast tissue penetration and low toxicity. They can be specifically labelled with various reporters for molecular recognition. Up to now, many of the published aptamer-nanoparticle assays or sensors for cancer detection are based on a handful of aptamers that are specific for T-cell leukaemia and B-cell lymphoma. This panel of DNA aptamers was selected directly from cancer cells by Cell-SELEX [26] for the recognition of molecular differences among leukaemia patient samples [27]. The generated aptamers enable differentiation between acute lymphoblastic T-cell leukaemia (CCRF-CEM cells [28]), and B-cell lymphoma (Ramos cells, human Burkitt’s lymphoma [29, 30] and Toledo cells [27]). All of the aptamers show high specificity for their respective target cells and equilibrium dissociation constants in the nanomolar to subnanomolar range.
Gold nanoparticles have been modified with these aptamers for use as targeted probes for imaging and studying lymphoma or leukaemia cancer cells [31–33].
One especially user-friendly application, which enables cancer detection very easily and in the shortest time with a strip-based assay [34], is worth spotlighting. A thiolated aptamer specific for Ramos cells was immobilised on gold nanoparticles and an additional Ramos aptamer (biotinylated) on the strip’s test zone. When the sample solution containing Ramos cells was applied to the sample pad, the solution migrated by capillary action past the conjugate pad and then rehydrated the aptamer conjugated gold nanoparticles. The Ramos cells interacted first with the modified nanoparticles and then continued migrating along the strip. When reaching the test zone, they were captured by a second reaction between the Ramos cells and the immobilised biotinylated aptamers. This is visualised as a characteristic red band due to the accumulation of gold nanoparticles in the test zone. Under optimal conditions, the test strip was capable of detecting a minimum of 4,000 Ramos cells by visual judgment only, without instrumentation. If a portable strip reader was used, a minimum of 800 cells were detectable. The measurements in buffer solution could be made within 15 minutes. The feasibility of the test system for the detection of cancer cells in biological fluids was evaluated and a successful determination of Ramos cells in human blood could be shown.
Chen et al. [35] produced an aptamer-conjugated fluorescence resonance energy transfer (FRET) nanoparticle assay that performs simultaneous multiplexed monitoring of cancer cells, in this case T-cell leukaemia and B-cell lymphoma cells. The authors firstly tuned the FRET-mediated emission signature by changing the doping ratio of three different dyes such that the nanoparticles would exhibit multiple colours upon excitation with a single wavelength. These FRET nanoparticles were then modified with a few aptamers specific for different cancer cell lines (Toledo, CCRF-CEM and Ramos cells). Fluorescent imaging (fig. 2) and flow cytometry were used for cellular detection, and demonstrated the selectivity and sensitivity of the method. Previously, Herr et al. [36] used similar dye-doped silica nanoparticles combined with aptamer-conjugated magnetic nanoparticles for selective collection and detection of acute leukaemia cells, and were able to separate and monitor these CCRF-CEM cells from mixed cell and whole blood samples.
Beside the use of the above-mentioned DNA aptamers for leukaemia cells, a few nanoparticle-based cancer detection assays have been described. For example, Lu et al. [37] reported a simple colourimetric and highly sensitive two-photon scattering assay for highly selective and sensitive detection of breast cancer SK-BR-3 cell lines at the 100 cells/ml level. They used oval-shaped gold nanoparticles, which they conjugated with a monoclonal antibody for human epidermal growth factor receptor and breast cancer specific RNA aptamers.
RNA aptamers directed against prostate-specific membrane antigen (PSMA) were developed by Lupold et al. [38]. These aptamers are the first reported RNA aptamers selected to bind a tumour-associated membrane antigen and the first application of RNA aptamers to a prostate cancer specific cell marker. The set of aptamers is very specific and even able to discriminate different human prostate cancer cell lines. The dissociation constants of the PSMA aptamers are in a low nanomolar range. Walter et al. [39] modified gold nanoparticles with these aptamers and used them in tissue microarrays for the detection of PSMA in human prostate cancer tissue.
A DNA aptamer for epithelial tumour marker mucin 1 (MUC1) was selected by Ferreira et al. [40]. MUC1 is a glycoprotein expressed on most epithelial cell surfaces and present in a variety of malignant tumours. The selected aptamers were shown to detect MCF-7 breast cancer cells. Cheng et al. [41] reported an aptamer-based, quantitative detection protocol for MUC1 using a three-component DNA hybridisation system with quantum dot-labelling. The sensor was based on a construct of three specially designed DNA strands (quencher, quantum dot-labelled reporter and the MUC1 aptamer stem), which allowed a strong fluorescence in the absence of the analyte. In the presence of MUC1 peptides, the fluorescence intensity decreased as a result of the structure switch of the aptamer strand when binding MUC1. In this way, the quencher and fluorescence reporter were brought into close proximity, which led to the occurrence of fluorescence resonance energy transfer, FRET, between the quencher and quantum dot. The detection limit for MUC1 with this approach was at the nanomolar level, and a linear response could be established for the approximate range found in blood serum. The method offers the possibility of improvement in the early diagnosis of different types of epithelial cancers.
The most advanced aptamer in the cancer setting is AS1411 (formerly known as AGRO100) – an experimental anticancer drug that was not generated using the SELEX process [42]. AS1411 is being administered systemically in clinical trials. This aptamer is a member of a novel class of antiproliferative agents known as G-rich oligonucleotides (GROs). These are non-antisense, guanosine-rich phosphodiester oligodeoxynucleotides that form stable G-quadruplex structures. The biological activity of GROs results from their binding to specific cellular proteins. One important target protein of GROs has been previously identified as nucleolin, a multifunctional protein expressed at high levels by cancer cells [43]. Ko et al. [44] used AS1411 for the development of a two-colour visualisation system for specific biomarker targeting by different quantum dot nanoparticle probes labelled with an aptamer and a targeting peptide. The so-called fluorescence derby imaging used dual colour quantum dots conjugated by the AS1411 aptamer (targeting nucleolin) on one hand and the arginine-glycine-aspartic acid (targeting the integrin alpha(v)beta(3)) on the other. The simultaneous fluorescence imaging of the cellular distribution of nucleolin and integrin using quantum dots enabled easy monitoring of separate targets in cancer cells and in normal healthy cells. These results suggest the feasibility of concurrent visualisation of quantum dot-based multiple cancer biomarkers using small molecules such as aptamer or peptide ligands.
Hua et al. [45] developed a novel strategy for selective collection and detection of breast cancer cells by combining the two aptamers mentioned above. The MUC1 aptamer was covalently conjugated to magnetic beads to capture breast cancer cells through affinity interaction between the aptamer and MUC1 protein. The MUC1 protein is overexpressed on the cancer cell surface. The captured cells were detected by a specially constructed nano-bio-probe consisting of the nucleolin aptamer AS1411 and quantum dots. The quantum dots were homogeneously coated on the surfaces of monodispersed silica nanoparticles. The nano-bio-probe attached to the surface of the pathogenic cells through the affinity of the AS1411 aptamer to nucleolin, which is also overexpressed in MCF-7 breast cancer cells. Photoluminescence and square-wave voltammetric assays were used for quantification, and detection limits of 201 cells/ml and 85 cells/ml, respectively, could be achieved. Simultaneous usage of the two aptamers as recognition elements improved selectivity.
Aptamers combined with nanoparticles hold great potential for effective detection of cancer even in the early stages of the disease. However, there are only a few aptamers available for different types of cancer cells currently. By development of further aptamers specific for cancer cells and markers, it will be possible to expand this field enormously.
Aptamers were first introduced as imaging probes for in-vivo studies in 1997, when Charlton et al. [46] used an aptamer selected against human neutrophil elastase labelled with a metastabile isotope (technecium-99; 99Tc) for imaging inflammation processes in a rat model. This aptamer showed a higher signal-to-background ratio than its antibody counterpart, demonstrating the potential applications of aptamers as imaging probes for in-vivo studies. Taking advantage of rapidly expanding nanobiotechnology-based developments, aptamer-nanoparticle conjugation forms the basis of a new chemical and biological strategy for in-vivo imaging. Because of their small size, nanoparticles can interact readily with biomolecules both on the surface and the cells. When conjugated with biomolecular affinity ligands, such as aptamers, they are considered to be a revolutionary approach for detection of various diseases that can be combined with directed therapy strategies.
Targeted metallic nanoparticles modified with aptamers as targeting agents have shown potential as a platform for development of molecular-specific contrast agents. Javier et al. [47] investigated the development of aptamer-based gold nanoparticles as contrast agents. The group devised a novel conjugation approach with an extended aptamer design where the extension was complementary to an oligonucleotide sequence attached to the surface of the gold nanoparticles. This conjugation approach was used to create a contrast agent designed to detect PSMA by obtaining reflectance images of PSMA-positive and PSMA-negative cell lines treated with the anti-PSMA aptamer-gold conjugates. Recently, Kim et al. [48] presented a multifunctional drug-loaded aptamer-gold nanoparticle bioconjugate for combined computed tomography (CT) imaging and therapy of prostate cancer. The surface of gold nanoparticles was functionalised with a RNA aptamer that binds to PSMA [38]. In this way, a targeted molecular CT imaging system capable of specific imaging of prostate cancer cells that express the PSMA protein was established.
To overcome limitations in targeted cell labelling related to molecule size and instability of the detection molecules, Zhou et al. [49] generated a new type of sub-10 nm multifunctional nanomaterial. They used the CCRF-CEM aptamer [28] and a dendrimer that additionally had the capacity to carry multiple fluorophore molecules. The hybrid DNA aptamer-dendrimer nanomaterial was used for labelling acute leukaemia cells. The results of binding studies with flow cytometry and fluorescence imaging microscopy showed high binding affinity and specificity of the constructed nanomaterial. Owing to the very small size of the created aptamer-dendrimers, the authors assume a principal applicability as contrast agents for specific in-vivo cancer imaging.
Most of these promising studies have been performed on the basis of in-vitro cell assays.
The in-vivo functionality of these novel multifunctional nanoparticles has to be proved by further studies. Nevertheless, in-vivo effects of aptamer-nanoparticles used for cancer visualisation in mouse models have recently been described [50]. The AS1411 aptamer specific to the nucleolin protein was conjugated to cobalt-ferrite nanoparticles surrounded by fluorescent rhodamine within a silica shell and to gallium-67 (67Ga). These multimodal nanoparticles were administered by intravenous injection into tumour-bearing nude mice and their biodistribution was analysed (fig. 3). The conjugates showed rapid blood clearance and accumulation in the tumour site, detected with scintigraphic images and magnetic resonance imaging. Furthermore, accumulation of the conjugate was corroborated by fluorescence imaging of the tumour after organ extraction. However, the conjugate was also shown to accumulate nonspecifically in liver and intestine, which may be a result of the size of the particles. Accumulation of nanoparticles in the liver is a widespread phenomenon and a problem independent of aptamers. To circumvent this problem, not only the influence of particle material, charge, and size, but also surface modifications are currently being studied [51]. One can foresee the promise of this technology once particle size can be optimised to minimise nonspecific uptake.
Apart from their use as a sensing platform for bioanalysis discussed above, aptamer-nanomaterial conjugates have also been applied in targeted drug delivery when used as carriers bound with cargo molecules, such as drugs or functional proteins. Cargos can be loaded onto nanoparticles either covalently or noncovalently. Covalent modifications of cargoes are generally achieved through standard gold-thiol chemistry, peptide bond formation or similar methods. For noncovalent introduction of cargo molecules, electrostatic adsorption and hydrophobic interaction schemes are commonly employed. Regardless of the kind of cargo loading, conjugated nanoparticles are very appropriate for in-vivoapplications because of their biocompatibility [52].
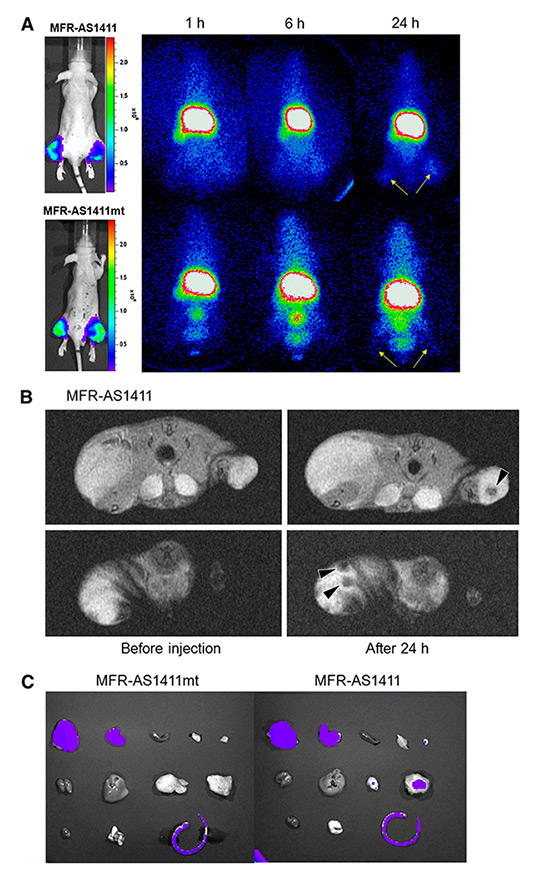
Figure 3
In vivo multimodal cancer targeting and imaging using MFRAS1411 particles. (A) MFR-AS1411 particles were intravenously injected into tumour-bearing mice and radionuclide images were acquired at 1, 6 and 24 h after injection. Scintigraphic images of C6 tumours in mice that received MFRAS1411 showed that C6 tumours had accumulated MFR-AS1411 at 24 h after injection but did not accumulate MFRAS1411mt (n 5 3). Tumour growth patterns were followed using bioluminescence signals acquired from luciferase-expressing C6 cells. (B) Magnetic resonance images of tumour-bearing mice before and after injection of MFR-AS1411 were acquired. Dark signal intensities at tumour sites were detected in MFR-AS1411–injected mice (arrowhead). (C) Tumours were isolated and their fluorescence verified using IVIS200 system. Fluorescence signal at tumour site injected with MFRAS1411 was detected, compared with tumours injected with MFR-AS1411mt. Isolated organs in order from upper left to lower right were intestine, liver, spleen, muscle, fat, kidney, stomach, right tumour, left tumour, heart, lung, and tail. Reprinted by permission of the Society of Nuclear Medicine from [50].
Farokhzad and co-workers have extensively studied the use of the anti-PSMA A10 RNA aptamer, and its truncated version A10-3, to target nanoparticles [24]. In a pioneering nanoparticle-targeting study, they demonstrated that the A10-3 aptamer can be used to target poly(lactic acid)-blockpolyethylene glycol copolymer nanoparticles to PSMA positive prostate cancer cells. The resulting nanomaterial showed a 77-fold increase in binding to PSMA expressing prostate cancer cells in comparison with untargeted nanoparticles [53]. The A10-3 aptamer was again used to target modified poly(D,L-lactic-co-glycolic acid) (PLGA) nanoparticles to deliver docetaxol to prostate tumours in-vivo, where complete tumour regression was found in five out of seven mice after a single intratumoural injection. Moreover, all of the treated animals survived a 109-day study [54]. In a next step, the group optimised their protocol for production of PEGylated PLGA nanoparticles and conjugated the resulting nanoparticles to the A10-3 aptamer and to docetaxel (and the related 14C-paclitaxel). After systemic administration, the delivery of these nanoparticles conjugates to tumours was enhanced 3.7-fold when compared with the nontargeted particles. Biodistribution patterns to the heart, lungs and kidneys of the treated mice did not show substantial accumulation of nanoparticles. However, the presence of high aptamer surface density led to an increase in nanoparticle accumulation in liver and spleen. This was likely to be due to aptamer masking the PEG layers on the surfaces of the nanoparticles and compromising the nanoconjugates antibiofouling properties in vivo. Thus, in engineering targeted nanoparticles, it is necessary to balance the tumour-targeting ligand surface density and the antibiofouling surface properties [55, 56].
In recent work, the Farokhzad group used a similar polymeric formulation to deliver cisplatin to PSMA expressing tumours by means of aptamer-functionalised Pt(IV) prodrug-PLGA-PEG nanoparticles. A dosage of 0.3 mg/kg of aptamer-targeted cisplatin nanoparticles was found to be more efficacious than a 1 mg/kg dosage of free cisplatin [57]. Furthermore, the same group demonstrated the feasibility of their systems for multiple drug therapy: targeted dual-drug combination based on nanoparticles with hydrophobic docetaxel and hydrophilic Pt(IV) drug. Superior efficiency over single-drug nanoparticle analogues or nontargeted nanoparticles could be demonstrated [58]. In an additional study, the anti-PSMA A10 aptamer was conjugated with superparamagnetic iron oxide nanoparticles and with a doxorubicin cargo, with aim of a dual function as combined prostate cancer imaging (with magnetic resonance) and therapy. The in-vitro cytotoxicity assay showed that the nanoparticle-mediated doxorubicin delivery and the delivery of free doxorubicin are equally potent against PSMA-positive cancer cells. More importantly, treatment with the aptamer-functionalised nanoparticles killed 47.5% of the PSMA-positive cells versus 22.8% of the PSMA-negative cells [59].
Aravind et al. [60] constructed anticancer drug-loaded lipid–polymer combinational hybrid nanoparticles. These nanoparticles were functionalised with AS1411 antinucleolin aptamers for site-specific targeting against tumour cells that overexpress nucleolin receptors. Cytotoxicity studies were carried out in two different cancer cell lines (breast cancer MCF-7 cells and human gliosarcoma GI-1 cells). Drug-loading studies indicated that with the same drug load, the aptamer-targeted nanoparticles show an enhanced cancer killing effect compared with the corresponding nontargeted nanoparticles. In addition, the lipid-polymer combinational nanoparticles exhibited high encapsulation efficiency and superior sustained drug release than the drug loaded in plain polymer nanoparticles.
The results of in-vitro imaging of cancer cells [41, 44] indicated that aptamer-conjugated quantum dots have the potential to be useful in imaging and protein expression profiling of living cells and fixed tissue, as well as in-vivo studies and drug delivery, although their use for the latter application may be limited by the relatively high cytotoxicity of quantum dots [61, 62]. However, there are intentions to use this material in vivo.For example, Savla et al. [63] attempted to apply the MUC1 aptamers [40] to target quantum dots with a doxorubicin cargo for imaging and for the chemotherapy of ovarian cancer. Doxorubicin was attached to quantum dots via a pH-sensitive hydrazone bond in order to provide stability of the complex in the systemic circulation and drug release in the acidic environment inside cancer cells. In a mouse model with human ovarian cancer xenografts, more MUC1 aptamer quantum dots accumulated in the tumours when compared with nonmodified quantum dots. Ex-vivo analysis of organs confirmed higher uptake in the tumour and lower uptake in other organs. The data obtained demonstrated the high potential of targeted quantum dot conjugates in the treatment of cancer. Besides other carriers, such as micelles and nanogels, aptamer-based drug delivery via nanoparticles promises to represent a new trend in specific therapeutic applications. It may contribute to the development of the next generation of nano-scale diagnostic and therapeutic modalities.
1 Andy Ellington’s Blog [Internet]. Austin: Andrew Ellington. 2010 July – . On Aptamers; 2011 March 6 [cited 2012 Dec 20]; [about three screens]. Available from: http://ellingtonlab.org/blog/2011/03/06/on-aptamers/
2 Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–22.
3 Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10.
4 Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287(5454):820–5.
5 Ozalp VC, Eyidogan F, Oktem HA. Aptamer-gated nanoparticles for smart drug delivery. Pharmaceuticals. 2011;4(2011):1137–57.
6 Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263(5152):1425–9.
7 Stoltenburg R, Reinemann C, Strehlitz B. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24(4):381–403.
8 James W. Aptamers. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. Chichester: John Wiley & Sons, Ltd.; 2000. P. 4848–71.
9 Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci U S A. 2003;100(26):15416–21.
10 Sefah K, Shangguan D, Xiong XL, O’Donoghue MB, Tan WH. Development of DNA aptamers using Cell-SELEX. Nature Protoc. 2010;5(6):1169–85.
11 Tucker CE, Chen LS, Judkins MB, Farmer JA, Gill SC, Drolet DW. Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J Chromatogr B. 1999;732(1):203–12.
12 Ni X, Castanares M, Mukherjee A, Lupold SE. Nucleic Acid Aptamers: Clinical Applications and Promising New Horizons. Curr Med Chem. 2011;18(27):4206–14.
13 Carlson B. Aptamers: The New Frontier In Drug Development? Biotechnol Healthc. 2007;(April):31–5.
14 Vallian S, Khazaei MR. Medical applications of aptamers. Res Pharm Sci. 2007;2(2):59–66.
15 Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10(3):282–9.
16 Green LS, Jellinek D, Bell C, Beebe LA, Feistner BD, Gill SC, et al. Nuclease-Resistant Nucleic-Acid Ligands to Vascular-Permeability Factor Vascular Endothelial Growth-Factor. Chem Biol. 1995;2(10):683–95.
17 Rhodes A, Deakin A, Spaull J, Coomber B, Aitken A, Life P, et al. The generation and characterization of antagonist RNA aptamers to human oncostatin M. J Biol Chem. 2000;275(37):28555–61.
18 Dougan H, Lyster DM, Vo CV, Stafford A, Weitz JI, Hobbs JB. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol. 2000;27(3):289–97.
19 Klussmann S. The Aptamer Handbook. Functional Oligonucleotides and Their Applications. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA; 2006.
20 Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21(2):74–81.
21 Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, et al. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32(19):5757–65.
22 Burke DH, Scates L, Andrews K, Gold L. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J Mol Biol. 1996;264(4):650–66.
23 Wilson C, Nix J, Szostak J. Functional requirements for specific ligand recognition by a biotin-binding RNA pseudoknot. Biochemistry. 1998;37(41):14410–9.
24 Xiao Z, Farokhzad OC. Aptamer-functionalized nanoparticles for medical applications: challenges and opportunities. ACS Nano. 2012;6(5):3670–6.
25 Phillips JA, Lopez-Colon D, Zhu Z, Xu Y, Tan W. Applications of aptamers in cancer cell biology. Anal Chim Acta. 2008;621(2):101–8.
26 Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res. 2010;43(1):48–57.
27 Shangguan DH, Cao ZHC, Li Y, Tan WH. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin Chem. 2007;53(6):1153–5.
28 Shangguan D, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103(32):11838–43.
29 Mallikaratchy P, Tang ZW, Kwame S, Meng L, Shangguan DH, Tan WH. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol Cell Proteomics. 2007;6(12):2230–8.
30 Tang ZW, Shangguan D, Wang KM, Shi H, Sefah K, Mallikratchy P, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79(13):4900–7.
31 Zhang K, Tan T, Fu JJ, Zheng T, Zhu JJ. A novel aptamer-based competition strategy for ultrasensitive electrochemical detection of leukemia cells. Analyst. 2013;138(21):6323–30.
32 Sheng W, Chen T, Tan W, Fan ZH. Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. ACS Nano. 2013;7(8):7067–76.
33 Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Gold nanoparticle-based colorimetric assay for the direct detection of cancerous cells. Anal Chem. 2008;80(4):1067–72.
34 Liu G, Mao X, Phillips JA, Xu H, Tan W, Zeng L. Aptamer-nanoparticle strip biosensor for sensitive detection of cancer cells. Anal Chem. 2009;81(24):10013–8.
35 Chen X, Estevez MC, Zhu Z, Huang YF, Chen Y, Wang L, et al. Using aptamer-conjugated fluorescence resonance energy transfer nanoparticles for multiplexed cancer cell monitoring. Anal Chem. 2009;81(16):7009–14.
36 Herr JK, Smith JE, Medley CD, Shangguan D, Tan W. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal Chem. 2006;78(9):2918–24.
37 Lu W, Arumugam SR, Senapati D, Singh AK, Arbneshi T, Khan SA, et al. Multifunctional oval-shaped gold-nanoparticle-based selective detection of breast cancer cells using simple colorimetric and highly sensitive two-photon scattering assay. ACS Nano. 2010;4(3):1739–49.
38 Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62(14):4029–33.
39 Walter JG, Petersen S, Stahl F, Scheper T, Barcikowski S. Laser ablation-based one-step generation and bio-functionalization of gold nanoparticles conjugated with aptamers. J Nanobiotechnology. 2010;8:21.
40 Ferreira CS, Matthews CS, Missailidis S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor Biol. 2006;27(6):289–301.
41 Cheng AK, Su H, Wang YA, Yu HZ. Aptamer-based detection of epithelial tumor marker mucin 1 with quantum dot-based fluorescence readout. Anal Chem. 2009;81(15):6130–9.
42 Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5(12):2957–62.
43 Watanabe T, Hirano K, Takahashi A, Yamaguchi K, Beppu M, Fujiki H, et al. Nucleolin on the cell surface as a new molecular target for gastric cancer treatment. Biol Pharm Bull. 2010;33(5):796–803.
44 Ko MH, Kim S, Kang WJ, Lee JH, Kang H, Moon SH, et al. In vitro derby imaging of cancer biomarkers using quantum dots. Small. 2009;5(10):1207–12.
45 Hua X, Zhou Z, Yuan L, Liu S. Selective collection and detection of MCF-7 breast cancer cells using aptamer-functionalized magnetic beads and quantum dots based nano-bio-probes. Anal Chim Acta. 2013;788:135–40.
46 Charlton J, Sennello J, Smith D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem Biol. 1997;4(11):809–16.
47 Javier DJ, Nitin N, Levy M, Ellington A, Richards-Kortum R. Aptamer-targeted gold nanoparticles as molecular-specific contrast agents for reflectance imaging. Bioconjug Chem. 2008;19(6):1309–12.
48 Kim D, Jeong YY, Jon S. A drug-loaded aptamer-gold nanoparticle bioconjugate for combined CT imaging and therapy of prostate cancer. ACS Nano. 2010;4(7):3689–96.
49 Zhou J, Soontornworajit B, Martin J, Sullenger BA, Gilboa E, Wang Y. A hybrid DNA aptamer-dendrimer nanomaterial for targeted cell labeling. Macromol Biosci. 2009;9(9):831–5.
50 Hwang do W, Ko HY, Lee JH, Kang H, Ryu SH, Song IC, et al. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. J Nucl Med. 2010;51(1):98–105.
51 Albanese A, Tang PS, Chan WC. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14:1–16.
52 Chen T, Shukoor MI, Chen Y, Yuan Q, Zhu Z, Zhao Z, et al. Aptamer-conjugated nanomaterials for bioanalysis and biotechnology applications. Nanoscale. 2011;3(2):546–56.
53 Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64(21):7668–72.
54 Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A. 2006;103(16):6315–20.
55 Cheng J, Teply BA, Sherifi I, Sung J, Luther G, Gu FX, et al. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28(5):869–76.
56 Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci U S A. 2008;105(7):2586–91.
57 Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci U S A. 2008;105(45):17356–61.
58 Kolishetti N, Dhar S, Valencia PM, Lin LQ, Karnik R, Lippard SJ, et al. Engineering of self-assembled nanoparticle platform for precisely controlled combination drug therapy. Proc Natl Acad Sci U S A. 2010;107(42):17939–44.
59 Wang AZ, Bagalkot V, Vasilliou CC, Gu F, Alexis F, Zhang L, et al. Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy. ChemMedChem. 2008;3(9):1311–5.
60 Aravind A, Jeyamohan P, Nair R, Veeranarayanan S, Nagaoka Y, Yoshida Y, et al. AS1411 aptamer tagged PLGA-lecithin-PEG nanoparticles for tumor cell targeting and drug delivery. Biotechnol Bioeng. 2012;109(11):2920–31.
61 Lopez-Colon D, Jimenez E, You M, Gulbakan B, Tan W. Aptamers: turning the spotlight on cells. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2011;3(3):328–40.
62 Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–44.
63 Savla R, Taratula O, Garbuzenko O, Minko T. Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin conjugate for imaging and treatment of cancer. J Control Release. 2011;153(1):16–22.
64 McCarthy JR, Weissleder R. Multifunctional magnetic nanoparticles for targeted imaging and therapy. Adv Drug Deliv Rev. 2008;60(11):1241–51.
Funding / potential competing interests: Federal Ministry of Education and Research, Germany (BMBF), Project funding within the Program NanoNature (Project NanoPharm 03X0094B)