Cryptosporidium spp. in drinking water
DOI: https://doi.org/10.4414/smw.2012.13683
Hans Peter
Füchslin, Thomas
Egli, Stefan
Kötzsch
Summary
In most rural areas and small communities in Switzerland the drinking water is supplied to the consumers after a minimum or even no treatment at all. However, it is just in these areas where drinking water from sources of agricultural activities can be contaminated by liquid manure and faeces of pasturing animals. The Swiss drinking water regulations are limited to the monitoring of E. coli, Enterococcus
spp. and total plate counts only. Hence, resistant pathogens, as for example Cryptosporidium spp., remain unnoticed. During a drinking water survey, which lasted from June 2003 to December 2004, water samples were collected from 3 selected rural sites in Switzerland. The drinking water was investigated for Cryptosporidium spp., E. coli, Enterococcus spp., Clostridium perfringens and other parameters. In all samples oocysts of Cryptosporidiumspp. were detected at elevated concentrations of up to 0.18 oocysts/l. Between 28% and 75% of the oocysts were found to be vital by the excystation method. Sampled oocysts collected from the three sites were subjected to genotyping and in one case the isolate was found to belong to the genotype of C. parvum. No evidence for increased incidents of diarrhoea in the past years was noted by local authorities.
Samples from rural sites in Switzerland
Introduction
Cryptosporidium is a coccidian protozoan parasite that can infect the intestine of warm blooded animals and of human beings causing severe diarrhoea. Cryptosporidia develop enduring forms, so called oocysts, which are spread in the environment by faecal contamination where they can survive for several months [1]. The disease is normally transmitted faecal-orally by smear infection but also by contaminated food and water [2]. The low infection dose of less than 30 oocysts [3] and its persistence against chlorination [4] makes this pathogen, even today, a challenge for drinking water distributors. To date 13 species in the genus Cryptosporidium are known that exhibit different human pathogenicity. The most virulent species for humans are Cryptosporidium hominis and Cryptosporidium parvum[5]. Whereas Cryptosporidium hominis is transmitted from humans to humans, Cryptosporidium parvum can be also transmitted from animals to humans and defines a so called zoonosis [6]. For a long time Cryptosporidium spp. received little attention. This changed with the numerous outbreaks of cryptosporidiosis due to contaminated drinking water were reported in industrialised countries such as the USA, GB, Japan and others [7]. Especially the outbreak in Milwaukee with estimated between 15,000–400,000 infected persons [8, 9] led to several investigations about this newly recognised pathogen and its risk to the health of drinking water consumers. Based on epidemiological studies of contaminated wells Haas and Rose defined an action level for Cryptosporidium in drinking water: “Whereas a Cryptosporidium concentration exceeding 0.1 oocysts/l can cause sporadically cryptosporidiosis cases in a population, concentrations over 0.3 oocysts/l will almost certainly cause a cryptosporidiosis outbreak [10].”
Nowadays, the regulations concerning Cryptosporidium in drinking water in the different countries are far from a common, generally accepted standard. In the USA the Environmental Protection Agency (US EPA) defines a tolerable yearly risk of 1 infected person in 10,000 from drinking water, which is often expressed as a risk of 10-4 per person and year [11]. According to the published dose-infection model [12] this complies with a Cryptosporidium concentration of only 0.0000327 oocysts/l [13]. Furthermore, particularly for Cryptosporidium a Maximum Contaminant Level Goal (MCL) is set at zero in the USA. The drinking water treatments with supposedly safe catchment zones must reach at least a 99% removal for Cryptosporidium [14] whereas public water systems, which use either surface water or ground water directly influenced by surface water, have to perform additional treatment to reduce the cryptosporidiosis infection risk [14]. In Great Britain water companies must use a process for treating the water to ensure that the average number of oocysts is less than 1 per 10 litres of water [15]. This threshold is based on the already mentioned observation that outbreaks of cryptosporidiosis have only occurred, when the concentrations of oocysts were in excess of 1 in 10 litres [16]. The European Union Drinking Water Guideline declares the bacterium Clostridium perfringens, whose spores can survive for extended periods of time in soil, as a persistent faecal indicator and a surrogate for Cryptosporidium spp.Therefore, in 100 ml of drinking water sample no C. perfringens should be detectable [17]. In Switzerland no regulation at all exists concerning Cryptosporidia in drinking water. The Swiss regulation states generally that drinking water should not contain microorganisms at a concentration harmful to human health [18].
The detection of Cryptosporidia spp. is time-consuming and expensive. Therefore, drinking water is not regularly checked for this pathogen. The officially compulsory microbiological drinking water tests in Switzerland are limited to the detection of E. coli and enterococci, and the heterotrophic plate count [19]. While E. coli dies relatively quickly in the environment, the oocysts of Cryptosporidium remain infectious for weeks to months. In contrast to E. coli, Cryptosporidium also survives in chlorinated drinking water. Therefore, one has to take into account that water that fulfills the quality criteria for drinking water can still contain persistent pathogens such as Cryptosporidium [20].
In drinking water distribution systems that include an initial treatment of the raw water no Cryptosporidium in the drinking water are expected in Switzerland. However, in agricultural areas the water from the water catchment zones can come into contact with grazing animal’s excrements and liquid manure. Animals that are infected with cryptosporidia excrete infectious microorganism that can infiltrate the groundwater. This water is usually not treated in any way and, therefore, the aim of this study was to estimate the risk for a Cryptosporidium infection through the consumption of drinking water in agricultural areas. A drinking water survey was performed from June 2003 to November 2004 analysing water samples from three rural sites in Switzerland. These three sites were selected out of 15 sites because of their elevated oocyst concentrations determined in a previous study [21]. The drinking water was investigated for Cryptosporidium spp. and bacterial faecal indicators such as E. coli, Enterococcusspp., C. perfringens and other parameters. Besides determining their concentration, the vitality of oocysts was assessed by the excystation method. Furthermore, the sampled oocysts were subjected to genotyping because different Cryptosporidiumspp. exhibit different pathogenicity.
Material and methods
Description of the three sampling sites
In 2003 a drinking water survey was carried out which included 15 rural sites in Switzerland [21] where drinking water was investigated for Cryptosporidium spp., E. coli Enterococcus, Clostridium perfringens and other parameters. Three of these sites exhibited increased oocyst concentrations and they were therefore investigated again. With the local authorities it was decided to publish the results anonymously; nevertheless, the general characteristics of the three sampling sites are described below.
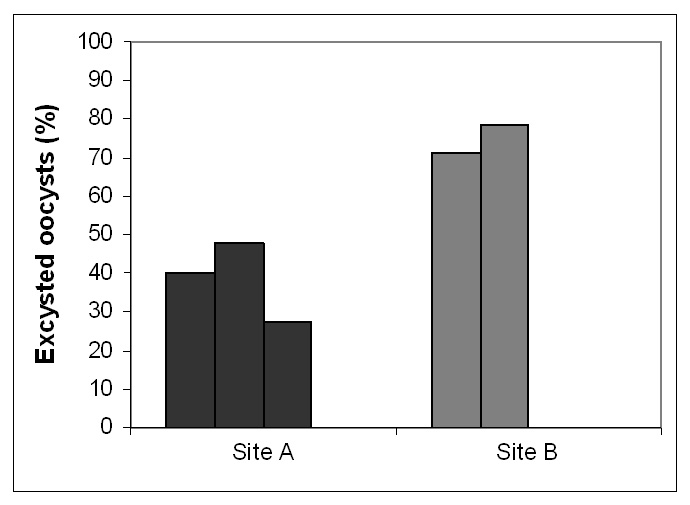
Figure 1
Viability of the oocysts in the drinking water samples determined by the excystation method. In the case of sites A 32, 42 and 125, in the case of sites B 11 and 12 oocysts, respectively, were analysed. Third sample of site B and samples from site C did not contain sufficient oocysts for a representive excystation testing.
Site A: Alpine village
Samples were collected from the drinking water distribution system of a village of approximately 500 habitants situated in the Swiss Alps. The tap water originated from a well and was neither chlorinated nor treated in any other way. It was suspected that the drinking water was contaminated by grazing cattle kept in the drinking water catchment zone.
Site B: Alpine camp
The second sampling site was the distribution network of a camp in the Swiss Alps. This camp has the capacity for several hundred people and has an independent well water supply. The drinking water was chlorinated after the first sampling campaign where faecal contamination was detected, probably caused by grazing cattle and sheep in the drinking water catchment zone.
Site C: Mittelland town in Switzerland
From the drinking water distribution system of a small town (around 8,000 habitants) samples were taken from the public water distribution network. The water was neither chlorinated nor treated in any other way. Cryptosporidium oocysts may originate from agricultural activities in the catchment zone from either pasturing animals or organic fertilisers such as manure.
The local authorities were informed about the results of this study and the detection of Cryptosporidium
sp. in their drinking water and if necessary measures were taken to improve its quality.
Sampling procedure
Water samples for bacterial analysis were collected in sterile 500 ml PET-bottles (Huber & CO. AG, Basel, Switzerland), which contained natrium thiosulfate in order to neutralise residual chlorine. For Cryptosporidium analysis between 100 to 1000 litres tap water were filtered through a Filta-Max® filter (IDEXX, Westbrook, USA) with a pore size of one µm. Samples were transported to the laboratory within less than 8 hours, protected from light and kept cool (12 °C). Samples were stored at 4 °C and analysed within 24 hours.
Detection of Escherichia coli, Enterococcus spp. and Clostridium perfringens
100 ml of sample were filtreed through a 0.45 µm membrane filter (Millipore, Bedford, USA). E. coli, Enterococcus
spp. and total plate counts were determined according to Swiss regulations [22], and C. perfringensaccording to the EU drinking water regulations [17]. Positive and negative controls were included.
Detection of Cryptosporidium spp.
Concentration of oocysts of Cryptosporidium spp. were determined according to method 1623, which is recommended by the Environmental Protection Agency [23]. The oocysts were detached from the Filta-Max® filter and further enriched and purified by Dynabeads® anti-Cryptosporidium (Invitrogen, Carlsbad, USA). The oocysts were stained with specific surface antibodies (Waterborne, New Orleans, USA) and finally enumerated by fluorescence microscopy. Loss of oocysts was not corrected for. Quality control of the method was performed with a commercial kit, Aqua-GloTM G/C Direct (Waterborne), obtaining a recovery of 29.75 ± 3.50% for Cryptosporidium oocysts.
Determination of viability of oocysts
In vitro excystation was performed according to a published protocol [24], in short: an aliquot of suspended bead oocyst complex (50 µl) was mixed with 50 µl of acidified Hank’s Buffered Salt Solution (HBSS); pH = 2.5, and incubated at 37 °C; after 30 minutes 50 µl of 3-times excystation medium (4.5% (wt/vol) tauroglycholic acid sodium salt, 1.5% bovine trypsine (2.2 U/µg), pH adjusted to 7.7 with NaHCO3) was added and the samples were further inoculated at 37 °C for at least 3 hours. The oocysts were washed with 0.22 µm-filtered PBS and later stained with fluorescent surface-specific antibodies (Waterborne, New Orleans, USA). The suspension was membrane-filtreed and finally the ratio of the excysted oocysts to the total number of oocysts was determined under the fluorescence microscope.
Extraction of DNA
A part of the immunomagnetically enriched and purified oocysts were further processed in order to extract their DNA for genotyping as follows: three freeze-thaw cycles in liquid nitrogen and a hot water-bath (96 °C) for 1 minute, each, were followed by proteinase K digestion at 56 °C for 4 hours. DNA was isolated using spin columns (DNA mini kit, Qiagen, Basel, Switzerland) in 200 µl of elution buffer according to the manufacturer’s instructions. DNA samples were frozen immediately at –20 °C.
Nested PCR for cryptosporidium
Nested PCR was done according to a published protocol [25]. In short: outer primers WR494F (TGA GTK AAG TAT AAA CCC CTT TAC) and AWA 1206R (CTC CAC CAA CTA AGA ACG GCC) were used amplifying a product of approximately 760 bp of the 18S rRNA – followed by the application of inner primers CBD-DiagF (AAG CTC GTA GTT GGA TTT CTG) and PW99R (TAA GGA ACA ACC TCC AAT CTC), which is a slightly modified AWA995 reverse primer. The product of approximately 420 bps contains a variable region suitable for genotyping Cryptosporidium
spp. PCR was done with 20 µl of the DNA extract (see above). The final concentration in the reaction mixture were 50 mM KCl, 20 mM Tris-HCl, pH 8.4, 2.5 mM MgCl2, 0.5% Tween 20, 400 ng/µl BSA, 200 µM dNTPs each, 1 µM of the primers, uracyl DNA glycosylase (UDG) and water to a final volume of 50 µl. Amplification was performed in a Genius Thermo Cycler (Techne, Cambridge, England) after incubating the reaction mixture for 10 min at 37 °C. Addition of Taq DNA polymerase (Sigma-Aldrich, St. Louis, USA) at 1.25 U per reaction was followed by an initial denaturation step at 94 °C for 10 min, 39 cycles at 94 °C for 30 sec, 58 °C for 40 sec, and 72 °C for 40 sec and a final extension step of 72 °C for 10 min. In the nested PCR, 1 µl of the first PCR array mixture was reamplified. Positive and negative controls were run in between every sample.
Detection and sequencing of PCR products
Reaction products were detected on agarose gels (1.5%) after staining with ethidium bromide (0.5 µg/ml) by visualisation with UV light, and their size was estimated by comparison with a sizemarker. A detection limit of 8 oocysts was experimentally defined in internal control experiments. Amplicons of the expected length of approximately 420 bp were purified by QIAamp DNA Mini Kit (Qiagen, Venlo, The Netherlands) and sent to a private company for sequencing in both directions (Microsynth, Balgach, Switzerland).
Sequence analysis
Only the part of the sequence that was confirmed by amplified fragments in both directions was used for further analysis. Sequence alignments for phylogenetic analysis were performed using MEGA version 3.1 [26]. After manual adjustment of the sequence with BLAST 2 SEQUENCES [27] and alignment by ClustalW [28] phylogenetic trees were constructed with neighbour joining methods using the Kimura 2-parameter model. Bootstrapping with 500 pseudoreplicates assessed the reliability of the tree. Additional Cryptosporidium 18S rDNA sequences were obtained from GenBank: C. felis (AF108862), C. hominis (DQ286403), C. meleagridis (AF180339), C. muris (AF093496.1), C. muskratgenotype (AY120904), C. parvum (AY030088), C. parvum (AF297513), C. parvum (AF030087), C. parvum (AF115377), C. parvum “pig genotype” (AF108861), C. wrairi (AF115378), C. serpentis (AF093499), C. sp. 5538 “C. muskrat genotype II” (AY545538). Perkinsus marinus(X75762) was used as an outgroup.
Nucleotide sequence accession numbers
The nucleotide sequence of the 18S rRNA gene of the Cryptosporidium sp.collected at drinking water sample site A has been deposited in GenBank under accession number EU263615.
Results
Oocysts concentrations at selected sites
Samples were taken from the three sampling sites at three different times (table 1). In the case of the alpine village (site A) all three samples had approximately the same oocyst concentration, whereas the oocyst concentrations in samples from site B varied strongly. The highest oocyst concentration was measured in the second sample after heavy rainfall. Due to chlorination no bacterial faecal indicators was found in this sample. Only the first sample of the town of the Swiss Mittelland had an elevated oocyst concentration whereas their concentrations in the second and third sample were close to the detection limit.
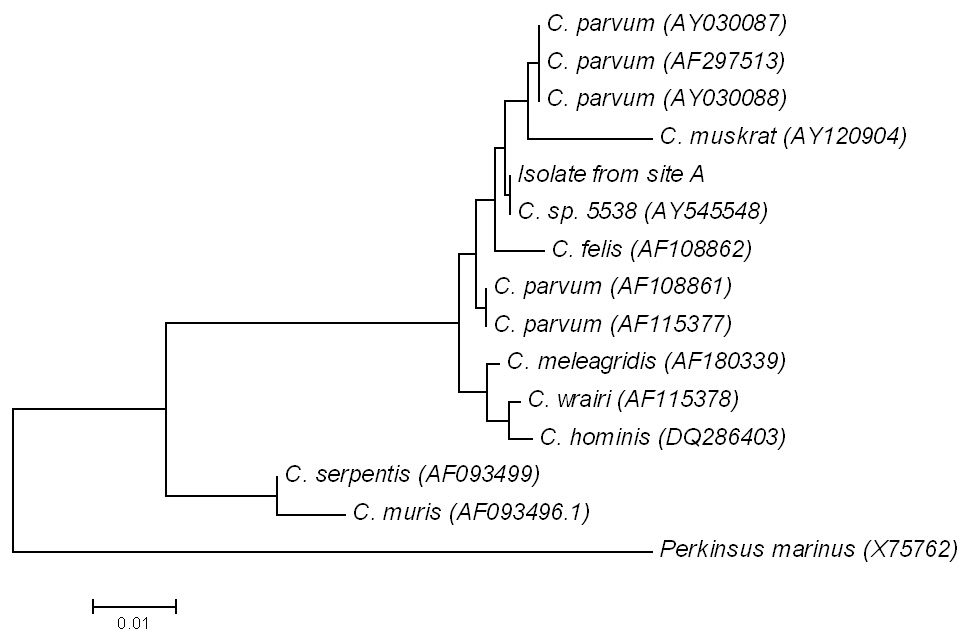
Figure 2
A neighbour-joining tree, based upon partial 18S rRNA gene sequence of the positive sample. For comparison, different Cryptosporidium strains and Perkinsus marinus X75762 as out-group were used.
In addition to the Cryptosporidium oocysts concentration, the presence of the faecal indicators E. coli, Enterococcus spp. and C. perfringens was measured. Furthermore, the heterotrophic plate count and nitrate concentration were determined (table 1). Only sporadic faecal indicators were detected. Therefore, no correlation between faecal indicators and oocyst concentration was observed. Also, neither nitrate concentration nor the heterotrophic plate count seem to correlate positively with oocyst concentration.
Viability
The viability of Cryptosporidium oocysts varied between 34% and 75% in the samples from site A and B (fig. 1). This is similar to values found in the literature, which assume that on average 40% of the oocysts in drinking water are viable [29]. The low number of oocysts in the sample from site C did not allow determination of their viability.
Genotyping
Only in the case of second sample of site A the nested PCR assays resulted in an amplification product. We can only speculate why in all other samples no amplification product could be found. This could be due to problems with extraction of the nucleic acid, inhibitors or oocyst concentration below detection limit. Just the part of the sequence (263-bp), which was confirmed by amplified fragments in both directions, was used for further analysis. The PCR-product of site A was sequenced and exhibited a very high 99% (262/263) similarity with different isolates of C. parvum(see fig. 2) and a 100% agreement with C. muskrat genotype IIAY545548(263/263). A phylogenetic tree was constructed with the sequence obtained from the drinking water sample of the site A and Genebank sequences (fig. 2). The isolate is located in cluster of different C. parvum strains. During sampling grazing cattle were present in the catchment zone of the village well. As C. muskrat infects only muskrats [30], which are not part of the fauna in the sampled region. Therefore, it is very likely that the detected Cryptosporidium strain most probably belongs to the genotype of C. parvum.
Risk assessment
The risk for the drinking water consumers of the site A was assessed based on the measured concentration, viability and genotype (table 2). The collected Cryptosporidium oocysts were viable and the amplified sequence is most probably belonging to the genotype of C. parvum suggesting a potential risk for the human health. The yearly risk of infection was calculated using the standard exponential model [12]. Assuming a daily consumption of 2 L of tap water with the average oocyst concentration of 0.154 oocysts/l resulted in a yearly infection risk of 40.8%. The assumed daily consumption of 2 L is very high. According to German study the average daily consumption of unboiled water is 0.17 L [31] and this assumption would lead to a yearly risk of 4.4%. We asked local authorities and the responsible physician of the district whether they observed increased numbers of diarrhoea cases. Both confirmed that no diarrhoeal outbreak was observed among the population in the alpine village. Located in the village is a home for elderly people, and also at this place, with around 70 inhabitants with a high proportion of potentially immuno-compromised consumers, no unusually high rate of cases of diarrhoea was observed.
|
Table 1: Summary of microbial and chemical analyses of samples from the three sampling sites. For bacterial analysis only one sample was taken in the first sampling. Later triplicate samples were collected. |
|
Place
|
Date
|
*Cryptosporidium
spp.
oocysts/l
|
E. coli
CFU/100 ml
|
Enterococcus spp.
CFU/100 ml
|
Cl. perfringens
CFU/100 ml
|
Heterotrophic
plate count
CFU/1 ml
|
NO3
–
concentration
mg/l
|
| Site A |
30.06.2003 |
0.146 |
|
5 |
|
1/6/1 |
15.46 |
| 23.09.2004 |
0.133 |
0/0/0 |
0/0/0 |
0/0/2 |
3/0/2 |
1.24 |
| 30.09.2004 |
0.183 |
0/0/0 |
0/0/0 |
0/1/2 |
3/3/1 |
1.23 |
| Site B |
26.08.2003 |
0.042 |
2 |
1 |
1 |
1/0/1 |
1.65 |
| 20.10.2004 |
0.109 |
0/0/0 |
0/0/0 |
0/0/0 |
3/3/5 |
2.63 |
| 16.11.2004 |
0.009 |
0/0/0 |
0/0/0 |
0/0/0 |
0/0/0 |
1.95 |
| Site C |
25.11.2003 |
0.038 |
|
|
|
6/11/7 |
26.55 |
| 09.09.2004 |
0.010 |
0/0/1 |
0/0/0 |
0/0/0 |
16/16/18 |
25.45 |
| 12.10.2004 |
0.010 |
0/0/0 |
0/0/0 |
0/0/0 |
2/2/4 |
25.27 |
| * Taking into account that in internal control experiments only 29.75 ± 3.50% of oocysts could be recovered, the reported oocysts concentrations have to be multiplied with the factor 3.3. |
|
Table 2: Risk assessment by comparison between measurements of the site A with facts of concern. |
| |
Results
|
Facts of concern
|
Possible consequences
|
|
Oocyst concentration
|
0.14–0.18/l |
>0.1 Ooc./l [10] |
Sporadically cryptosporidiosis cases or even outbreaks |
|
Oocystviability
|
28%–48% |
>0 |
Viability is a preliminary for infectivity |
|
Genotyping
|
According to our assessment the isolated strain is part ofC. parvumgenotype. |
C. parvum is a pathogenic strain for human |
Cryptosporidiosis among drinking water consumers possible |
Discussion
At three selected sites in Switzerland we detected Cryptosporidium oocysts in the concentration range between 0.04 and 0.18 oocysts/lin the drinking water. A number of reasons can cause the observed different oocysts concentrations of site A, B and C This includes the number of oocysts in the faeces of the pasture livestock, the amount of faeces distributed by pasturing animals, or the amount of manure distributed. Furthermore, weather conditions influence the behaviour of oocysts; for example, during heavy rainfall fresh faecal contaminants can infiltrate the soil quickly, whereas during dry weather faecal contaminants remain on the surface. Also soil structure has an impact on the flow velocity of the infiltrating water and the retention capacity for oocysts. Long residence times favour the natural elimination of microorganisms and viability of oocysts will decrease with increasing exposure to environmental stress factors [32]. Oocysts isolated from samples of the site A were viable and most probably members of the C. parvum genospecies. This is in agreement with the fact that within the drinking water catchment zone solid manure or faeces of pasturing cattle was observed during the sampling campaign. Young cattle are often suffering from cryptosporidiosis and have a prevalence of up to 16.8% in Switzerland [33]. These animals excrete several millions of C. parvum oocysts per day in faeces [34] and can be a significant source of infection for humans. This could be an explanation, why in contrast to other countries, where C. hominis is the main reason for Cryptosporidiuminfection for humans, C. parvum seems to be the main pathogen in Switzerland [35]. For example, in an epidemiological study of diarrhoea patients in Switzerland carried out by Fretz and co-workers all of the nine isolates from stool samples belonged to the genotype II of C. parvum and the zoonotic cycle was believed to play a major role [35]. This is supported by an earlier study where seven out of 13 isolates from HIV-infected patients belonged to the bovine C. parvum genotype [36]. In another Swiss study on children suffering from diarrhoea mainly C. hominis was detected (11 of 14 children, nine with history of travel), whereas the other children had the zoonotic genotype II of C. parvum(three children, one with travel history). All this implies cattle as an important source of autochthonous infections [37].
In general, cryptosporidiosis is of minor importance in Switzerland. The prevalence of diarrhoea cases, i.e., the percentage of the Cryptosporidium infected diarrhoea patients for the total population in Switzerland at a specific time, is only 0.2% [38]. Diarrhoeic children are more often infected with Cryptosporidium at a proportion of 4.6%–5.5% [37, 39, 40], and for AIDS patients the prevalence rises even to 11.8% [41]. Based on extrapolated clinical data an estimate of only 340 annual cases of cryptosporidiosis in Switzerland was calculated [38]. It is believed that consumption of raw diary products and oysters as well as travelling are the main risk factors whereas contaminated drinking water seems to be of minor importance [35, 38]. In several studies Cryptosporidium sp. oocysts were detected in Switzerland with highest reported concentration of 3.83 oocysts/l in surface water [25, 42–44], 1.6 oocysts/l in well water [42, 45], and 0.25 oocysts/lin tap water samples [42]. Despite this, no waterborne cryptosporidiosis have been reported in Switzerland up to now [25]. The reasons for the fact that no outbreaks were observed although viable oocyst concentrations of C. parvum are above the action level can be only speculated about. It is possible that the specific C. parvum strain present in grazing cattle in Switzerland is less pathogenic than normally assumed. Furthermore, the determined viability may not correlate with infectivity, i. e., that the oocyst lose their infectivity but remain viable. Another reason may be that local tap water consumers get immunised already in their childhood after a first infection with C. parvum oocysts and are more or less protected from further infections. Studies with human volunteers indicate that prior infection with Cryptosporidium offers a certain degree of protection against infection with low doses of oocysts [46]. Such a situation can be expected by a regular consumption of contaminated drinking water. Furthermore, it was shown in a study from northern Italy that local people had an increased serological response against Cryptosporidium oocysts caused by previous exposure to Cryptosporidium and explaining the observed lower risk for cryptosporidiosis outbreaks in this region [47]. But also simple other reasons have to be considered, such that people consume very little unboiled tap water, probably even less than 0.17 L per day [31], or that infected persons suffering from mild diarrhoea would not consult a doctor. Therefore, one cannot exclude that sporadic cryptosporidiosis or even outbreaks were missed.
The European Union Drinking Water Guideline [17] declares the spore-forming bacterium Clostridium perfringens as a persistent faecal indicator and surrogate for Cryptosporidiumspp. According to our results C. perfringens does not seem to be an appropriate indicator for C. parvum. Repeatedly, we detected Cryptosporidium oocyst in the absence of C. perfringens (table 1). This observation is also in agreement with other studies [21, 48]. No significant correlation with the C. parvum oocyst concentration was found for other chemical parameters such as nitrate or other microbial parameters. This suggests that there is presently no valid surrogate concept, that could replace the time-consuming and expensive direct detection for Cryptosporidium spp. Even if the detection of Cryptosporidium oocysts in the alpine region in this study may be of minor risk for human health, their presence in tap water proves that this drinking water was faecally contaminated. This is an alarming signal, also because, other pathogenic microorganism from liquid manure or faeces of pasturing animals such as Giardia spp., E. coli O157:H7, Salmonella spp., rotaviruses and other enteric microorganisms might contaminate the drinking water [49]. Especially after heavy rainfalls there is a danger that pathogens can temporarily break through into drinking water wells. Our study has shown that especially in rural regions the drinking water can be temporarily microbiologically contaminated. Nevertheless, there is currently no indication of waterborne cryptosporidiosis in Switzerland as reported by public health authorities [38].
Acknowledgements: We thank SPIEZ LABORATORY for financial support and Prof. Alexander Mathis of Institute of Parasitology, University of Zürich for his advice and help with PCR analysis and Genotyping for Cryptosporidium.
References
1 Carey C. Biology, persistence and detection of Cryptosporidium parvum and Cryptosporidium hominis oocyst. Water Res. 2004;38:818–62.
2 Juranek DD. Cryptosporidiosis: sources of infection and guidelines for prevention. Clin Infect Dis. 1995;21(Suppl 1):S57–61.
3 DuPont HL, Chappell CL, Sterling CR, Okhuysen PC, Rose JB, Jakubowski W. The infectivity of Cryptosporidium parvum in healthy volunteers. N Engl J Med. 1995;332(13):855–9.
4 Campbell IS, Tzipori S, Hutchinson G, AK W. Effect of disinfectants on survival of Cryptosporidium oocysts. Vet Rec. 1982;111:414–5.
5 Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17(1):72–97.
6 Peng MM, Xiao L, Freemaan AR, and al. e, Genetic polymorphism among Cryptosporidium parvum isolates: Evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–73.
7 Smith HV, Rose JB. Waterborne cryptosporidiosis: current status. Parasitol Today. 1998;14(1):14–22.
8 MacKenzie WR, Schell WL, Blair KA, Addiss DG, Peterson DE, Hoxie NJ, et al. Massive outbreak of waterborne Cryptosporidium infection in Milwaukee, Wisconsin: recurrence of illness and risk of secondary transmission. Clin Infect Dis. 1995;21(1):57–62.
9 Hunter PR, Syed Q. Community surveys of self-reported diarrhoea can dramatically overestimate the size of outbreaks of waterborne cryptosporidiosis. Water Sci Technol. 2001;43(12):27–30.
10 Haas CN, Rose JB. Developing an action level for Cryptosporidium. AWWA. 1995;87(9):81–3.
11 USEPA, National Primary Drinking Water Regulations: filtration, disinfection, turbidity, Giardia lamblia, viruses, Legionella, and heterotrophic bacteria; Final Rule, in Federal Register (40 CFR Parts 141 and 142). 1989.
12 Teunis P, Havelaar A. Risk assessment for protozoan parasites. Internat Biodeterior & Biodegr. 2002;50:185–93.
13 Metzler A, Tabisch A. Fakten und Spekulationen über die Kontamination der Umwelt mit Cryptosporidium-Oozysten. Gas, Wasser, Abwasser (GWA), 1998;78(1):32–6.
14 USEPA, National Primary Drinking Water Regulations: Long Term 2 Enhanced Surface Water Treatment Rule, in Federal Register. 2006, United States Environmental Protection Agency. p. 653–702.
15 HMSO, The Water Supply (Water Quality) (Amendment) Regulations 199, in Statutory Instrument. 1999, Her Majesty’s Stationery Office: London.
16 Haas CN. Epidemiology, microbiology, and risk assessment of waterborne pathogens including Cryptosporidium. J Food Prot. 2000;63(6):827–31.
17 European Communitiy Directive 98/83/EC, Council Directive 98/83/EC of 3 November 1998 on the quality of water intended for human consumption as amended by Regulation 1882/2003/EC. 1998(31998L0083 (32003R1882)).
18 LMG, Schweizerisches Bundesgesetz über Lebensmittel und Gebrauchsgegenstände (LMG), in 817.0. 1992.
19 HyV, Schweizerische Hygieneverordnung des EDI vom 23. November 2005 (HyV), in 817.024.1. 2005.
20 Smith HV, Smith PG. Parasitic protozoa in drinking water. Endeavour. 1990;14(2):74–9.
21 Füchslin HP, Beuret C, Egli T. Mikrobiologische Belastung des Trinkwassers in Trinkwasserfassungen ländlicher Regionen. Gas, Wasser, Abwasser (GWA), 2005;11:859–65.
22 SLMB, Schweizerisches Lebensmittelbuch. Schweizerische Eidgenossenschaft, 2000. Kapitel 56: Mikrobiologie.
23 USEPA, Method 1623: Cryptospordium and Giardia in water by filtration/IMS/FA. EPA, 1999. 821-R-99-006(Office of Water).
24 Wiedenmann A, Steuer S, Krüger P, Botzenhart K. A simple procedure for an exact evaluation of the sensitivity of the selective detection of viable Cryptosporidium oocysts by in vitro excystation and PCR. In: International Symposium on Waterborne Cryptosporidum Proceedings. 1997. Denver, Colorado, USA: AWWA.
25 Ward PI, Deplazes P, Regli W, Rinder H, Mathis A. Detection of eight Cryptosporidium genotypes in surface and waste waters in Europe. Parasitol. 2002;124(Pt 4):359–68.
26 Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutinary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–63.
27 Tatusova TA, Madden TL. Blast 2 sequences – a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–50.
28 Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80.
29 Teunis P, Havelaar A. Cryptosporidium in drinking water: evaluation of the ILSI/RSI Quantitative Risk Assessment Framework. 1999, Rijksinstituut voor Volksgezondheid en Milieu RIVM: Bilthoven
30 Zhou L, Fayer R, Trout JM, Ryan UM, Schaefer FW, 3rd, Xiao L. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl Environ Microbiol. 2004;70(12):7574–7.
31 Schneider AT, Eberhardt R, Hartmann S, Herwig A, Heseker H, Hünchen K, et al. Ergebnisse der Nationalen Verzehrsstudie (1985–1988) über die Lebensmittel- und Nährstoffaufnahme in der Bundesrepublik Deutschland, in Band XI der VERA-Schriftenreihe. 1995, Wissenschaftlicher Fachverlag Dr. Fleck: Niederkleen.
32 Auckenthaler A, Huggenberger P. Pathogene Mikroorganismen im Grund- und Trinkwasser. 2003, Basel: Birkhäuser Verlag.
33 Lentze T, Hofer D, Gottstein B, Gaillard C, Busato A. Prevalence an importance of endoparasites in calves raised in Swiss cow-calf farms. Deutsche tierärztliche Wochenschrift. 1999;106:275–81.
34 Nydam DV, Wade SE, Schaaf SL, Mohammed B. Number of Cryptosporidium parvum oocysts or Giardia spp. cysts shed by dairy calves after natural infection. Am J Vet Res. 2001;62(10):1612–5.
35 Fretz R, Svoboda P, Ryan UM, Thompson RC, Tanners M, Baumgartner A. Genotyping of Cryptosporidium spp. isolated from human stool samples in Switzerland. Epidemiol Infect. 2003;131(1):663–7.
36 Morgan U, Weber R, Xiao L, Sulaiman I, Thompson RC, Ndiritu W, et al. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. 2000;38(3):1180–3.
37 Gläser C, Grimm F, Mathis A, Weber MD, Nadal D, Deplazes P. Detection and molecular characterization of Cryptosporidium spp. isolated from diarrhic children in Switzerland. Pediatr Infect Dis J. 2004;23(4):1–3.
38 Baumgartner A, Marder HP, Munzinger J, Siegrist HH. Frequency of Cryptosporidium spp. as cause of human gastrointestinal disease in Switzerland and possible sources of infection. Schweiz Med Wochenschr. 2000;130(36):1252–8.
39 Essers B, Burnens AP, Lanfranchini FM, Somaruga SG, von Vigier RO, Schaad UB, et al. Acute community-acquired diarrhea requiring hospital admission in Swiss children. Clin Infect Dis. 2000;31(1):192–6.
40 Mäusezahl D, Egger M, Odermatt P, Tanner M. Clinical aspects and epidemiology of cryptosporidiosis in immunocompetent children. Schweiz Rundsch Med Prax. 1991;80:936–40.
41 Weber R, Ledergerber B, Zbinden R, Altwegg M, Pfyffer GE, Spycher M, et al. Enteric infections and diarrhea in human immunodeficiency virus-infected persons: prospective community-based cohort study – Swiss HIV Cohort Study. Arch Intern Med. 1999;159:1473–80.
42 Svoboda P, Ruchti S, Bissegger C, Tanner M. Occurence of Cryptosporidium spp. oocysts in surface, raw and drinking water samples. Mitt. Lebensm. Hyg. 1999;90(5):553–63.
43 Regli W. Improved methods for the isolation and detection of Giardia-cysts and Cryptosporidium-oocysts form surface and drinking water: flocculation and FACS, in Vetsuisse Faculty of Zurich. 1994, University of Zurich, Switzerland: Zurich.
44 Schweizer K. Modification and use of a method with FACS for the detection and quantification of Cryptosporidium oocysts in water, in Vetsuisse Faculty of Zurich. 1998, University of Zurich, Switzerland.
45 Auckenthaler A, Raso G, Huggenberger P. Particle transport in a karst aquifer: natural and artificial tracer experiments with bacteria, bacteriophages and microspheres. Water Sci Technol. 2002;46(3):131–8.
46 Chappell CL, Okhuysen PC, Sterling CR, Wang C, Jakubowski W, DuPont HL. Infectivity of Cryptosporidum parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am J Trop Med Hyg. 1999;60(1):157–64.
47 Frost FJ, Fea E, Gilli G, Biorci F, Muller TM, Craun GF, Calderon RL. Serological evidence of Cryptosporidium infections in southern Europe. Eur J Epidemiol. 2000;16(4):385–90.
48 Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol. 2005;71(6):3163–70.
49 Pell AN. Manure and microbes: public and animal health problem? J Dairy Sci. 1997;80(10):2673–81.

