HLA and non-HLA polymorphisms in renal transplantation
DOI: https://doi.org/10.4414/smw.2012.13668
Stéphanie
Laperrousaz, Jean-Marie
Tiercy, Jean
Villard, Sylvie
Ferrari-Lacraz
Summary
Despite progress made in the field of immunosuppression, graft rejection remains a major cause of morbidity and mortality of patients after solid organ transplantation. There are several genetic causes which could influence the outcome of renal transplantation. One of the main determining factors of success in renal transplantation is human leukocyte antigen (HLA) compatibility between donor and recipient, particularly at HLA-A, HLA-B and HLA-DR loci. HLA compatibility remains an essential immunological barrier, despite modern immunosuppressive treatments. There is also evidence that natural killer (NK) cell alloreactivity contributes to the immune response which modulates the outcome of renal transplantation. However, the clinical impact of combinations of KIR genes (family of NK cell receptors) and their HLA ligands in donor and recipient still remains to be clearly established. Furthermore, cytokines are involved in the immune reaction against the renal transplant, but the implication of the genetic polymorphism of cytokines is strongly debated. Therefore, while HLA compatibility remains a primordial component for any renal transplantation, it would be premature to use the two other genetic aspects as criteria for organ allocation and as prognostic factors.
Introduction
Solid organ transplantation between two genetically unrelated individuals induces a powerful immune response in the recipient leading to the rejection of the graft. This immunological reaction is largely determined by genetic factors. The strategies to inhibit this reaction include optimal genetic compatibility between donor and recipient and the use of immunosuppressive treatment to prevent rejection. In addition to the classical human leukocyte antigen (HLA) system, other immunogenetic factors have emerged as being of significant interest in transplantation immunology. The present review summarises the current knowledge of the significance of HLA compatibility, focusing on the KIR receptors of natural killer (NK) cells and on the genetic polymorphism of several cytokines (IL-6, IL-10, TNF-α and TGF-β). These non-HLA immunogenetic markers have been evaluated in a limited number of publications, mainly in regard to kidney transplantation.
The HLA polymorphism
The HLA genes are clustered on the short arm of chromosome 6 and code for the 3 HLA class I antigens (HLA-A, -B, -C) and the 3 HLA class II antigens (HLA-DR, -DQ, -DP) that are relevant to transplantation. The HLA genes constitute a multigenic system with a high degree of allelic polymorphism in human populations, with over 7,500 different alleles and over 5,458 expressed HLA antigens currently known (fig. 1).

Figure 1
MHC genes alleles. Short arm of chromosome 6 with representation of three HLA class I genes and three HLA class II genes. The latter have a different gene that encodes each α and β chain forming the final HLA class II molecule. These genes encode polymorphic proteins (over 5,400) known currently and are involved in the presentation of antigens to T cells. Moreover, the total number of alleles for the HLA loci is over 7,527, including silencing alleles (IMGT/HLA database).
An additional level of complexity exists in the HLA-DR sub-region. Whereas all individuals have a DRB1 gene that codes for alleles of the DR1-DR18 serotypes, about 90% of individuals have a second DRB gene: DRB3, DRB4 or DRB5. This second DRB locus codes for the much less polymophic serotypes DR52, DR53, and DR51 respectively.
Since HLA genes are co-dominantly expressed as cell surface glycoproteins, a heterozygous individual may express up to 14 different HLA antigens. The biological function of HLA molecules is to present peptide antigens to T lymphocytes. The HLA diversity is important in human populations as HLA polymorphism gives rise to a broader peptide antigen recognition and presentation to T cells, allowing an optimal adaptive immune response.
HLA polymorphism thus represents an important immunological barrier in solid organ transplantation, and the risk of acute/chronic rejection due to incompatible HLA antigens persists. The number of HLA disparities increases the risk of graft failure, and the better the recipient/donor HLA compatibility, the better the chances for a successful transplantation [1–3]. Due to their high allelic polymorphism, HLA molecules are potent inducers of the immune response and anti-HLA antibodies develop after exposure to allo-HLA antigens, typically after blood transfusion, pregnancy and previous transplantation [4]. The de novo development of these anti-HLA antibodies is a risk factor for graft rejection and the presence of preformed anti-HLA antibodies also represents a barrier to successful transplantation [5, 6].
Although the impact of HLA compatibility has been recognised for 2 decades (reviewed by Wujciak & Opelz) [7], the advances in immunosuppression protocols are such that rejection episodes are managed more efficiently, thus minimising the importance of HLA matching. A study based on the UNOS data reported that the impact of HLA compatibility had greatly diminished [8]. Consequently, some allocation programmes have gradually toned down the role of HLA matching from the algorithms used for prioritisations on the waiting list. The lower importance of HLA matching was also favoured in the early years of the 21st century with the advent of the microarray-based luminex technology for detecting donor-specific antibodies (DSA), thus allowing organ allocation around well characterised HLA specificities.
However, results of the large CTS study cohort (135,970 kidney transplants) clearly showed that HLA matching on graft survival rate has not lost its importance, and its significance is obvious when comparing the decades 1985–1994 and 1995–2004 [9]. Even when analysing the last 5 years of the study period separately (2000–2004), a significant correlation of graft survival with HLA matching was disclosed [9]. An analysis of the SRTR data base (1988–2007) consisting of >15,000 re-transplant candidates revealed the negative effect of poor HLA matching on graft survival after the first transplantation associated with a significant increase in the development of anti-HLA antibodies (measured by panel reactive antibody (PRA)) proportional to increasing HLA mismatches. Only 10% of patients with 0 HLA-A and -B-mismatches became newly sensitised after graft loss compared to 37% (>30% PRA) in transplants with a greater extent of HLA mismatches [10].
In addition to the classical HLA-A, -B and -DR antigens, the role of HLA-C and -DQ antigens in terms of graft survival or sensitisation is now documented [11, 12]. Analysis of the immunogenicity of incompatible HLA-A, -B antigens in terms of the numbers of amino acid residue mismatches (epitopes mismatches) is associated with better transplant outcome than conventional matching based on HLA typing by serology [13]. Although anti-DP antibodies are frequently detected in sensitised patients, their impact on transplant outcome is still not clear and needs to be further evaluated.
In the context of kidney transplantation from live donors, donor age and HLA matching have recently been shown to be independent donor-related risk factors associated with both decreased patient and graft survival [14].
Based on a better understanding of anti-HLA immunisation, Duquesnoy [15, 16] developed a new computer algorithm, HLAMatchmaker, making it possible to assess HLA mismatch acceptability based on the epitopes recognised by anti-HLA antibodies. These epitopes are characterised by crucial amino acid residues (so-called “eplets”) that dominate in antigen-antibody binding. HLA matchmaker represents a useful tool for determining histocompatibility at the epitope level that goes beyond the classical HLA-A, -B, -DR matching algorithm.
Natural killer cells and polymorphism of KIR receptors
Natural killer (NK) cells
“Natural killer” cells are large lymphocytes of the innate immune system [17–19] that represent 5–25% of mononuclear circulating cells. They establish the first line of defence of the body against intracellular infections and tumoural cells [17–19]. The two main effective functions of NK cells are lysis of the target cells by cytotoxic granules and the secretion of pro-inflammatory cytokines (ex: IFN-γ and TNF-α) [17–19]. NK cells possess various receptors on their surface. Some receptors issue activating signals and others issue inhibitory signals (fig. 2) [19]. The following section is devoted exclusively to “killer-cell immunoglobulin-like receptors” (KIRs).
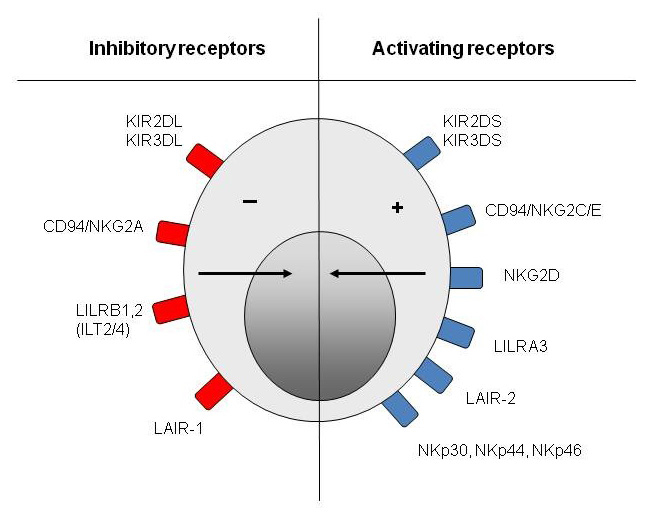
Figure 2
Natural killer cell receptors. Natural killer cells express activating and inhibitory receptors belonging to specific families of receptors like the KIR family.

Figure 3
Activation and inhibition of natural killer cells. The KIR protein can be divided into activating KIR (i.e KIR2DS [S for short]) or inhibitory KIR (i.e. KIR2DL [L for long]). The activating and inhibition signal is mediated by the short or the long tail of the KIR receptor after the binding with the HLA-C ligand. ITIM and ITAM motifs drive the inhibitory and activating signal respectively by a phosphorylation process.

Figure 4
The regulation of natural killer cell responses. The natural killer cell response is dependent on the balance between activating, inhibitory receptors and the expression of their ligand on the surface of target cells.
The absence of ligand is characterised by the absence of signalling (1). The expression of ligand (HLA-C1 in blue as a prototypical example) for inhibitory receptor (iKIR in red) is characterised by an inhibitory signal (2). The absence of ligand for inhibitory receptor in presence of activating receptor with their cognate ligand induce an activation of the NK cells, that is called the “missing self” (3). The balance of activating and inhibitory receptors in presence of their ligand will drive the state of NK cells activation (4). The presence of activating receptor with their ligand in absence of the specific ligand (HLA-C2 in grey as a prototypical example) for inhibitory receptor (iKIR in red) activates NK cells. Such situation is called the “missing ligand” (5).
KIR genes
The KIR genes are located in the region of the genome called “leukocyte-receptor complex” (LRC) on the chromosome 19 [19, 20]. The family of KIRs consists of fifteen genes that code for activating or inhibitory KIRs, and of two pseudo-genes which do not code for a functional protein [19]. In the LRC, the KIR genes are organised in the form of haplotypes which vary according to the number and the type of KIR genes present [19, 21]. In the human system, the segregation of these various haplotypes generates a large variety in numbers and types of KIR genes inherited by an individual. As for HLA genes, there is also the extensive polymorphism of KIR genes [19, 20] which influences their expression at the cell surface and their affinity to specific KIR ligands. A given NK cell expresses only a single part of the KIR genes present in its genome [18, 22]. The expression of a certain KIR gene is random. Consequently, each NK cell expresses its own repertoire of KIR receptors and this account for a great heterogeneity of NK cells within an individual [23, 24].
KIR proteins are cellular receptors which are either activating (aKIRs) or inhibitory (iKIRs) (fig. 2 and 3) [18, 19]. They consist of two or three immunoglobulin-like domains in their extracellular extremities which serve as the site of interaction for the specific ligand, of a transmembrane region and of a cytoplasmic region which is responsible for the signal transduction within the NK cell (fig. 3). KIR molecules formed by two or three immunoglobulin-like domains are named KIR2D or KIR3D. Furthermore, according to the length of the cytoplasmic tail which can be either long or short, the receptors are called KIR2DS (for short) or KIR2DL (for long) (fig. 3). Generally, a long cytoplasmic tail sends an inhibitory signal and a short cytoplasmic tail produces an activating signal inside the NK cell. There is, however, an exception for the KIR2DL4 receptor which can simultaneously generate activating and inhibitory signals [18, 19, 25, 26] ( http://www.ebi.ac.uk ).
KIR functions
aKIRs recognise a heterogeneous group of ligands that are expressed by cells undergoing stress (ex: intracellular infection, tumoural transformation) [18, 19]. They induce activation of NK cells. iKIRs link to HLA class I molecules which are expressed on all healthy nucleated cells [18, 19, 27]. Engagement of iKIRs inhibits the activation of NK cells.
Known combinations for the interaction between iKIRs and HLA class I are the following [18–20]:
– KIR2DL2/3 + HLA-C from group C1 (Cw1/3/7/8) carrying aspargin in position 80.
– KIR2DL1 + HLA-C from group C2 (Cw2/4/5/6/15) carrying lysin in position 80.
– KIR3DL1 + HLA-A/HLA-B with a Bw4 serological epitope.
– KIR3DL2 + HLA-A3 and HLA-A11.
The final integration of all KIR signals, generated by a variety of activating and inhibitory receptors at the time of interaction with the target cell, determines the behaviour/action of the NK cell [18, 19]. NK cells are capable of discriminating between a healthy cell which must be preserved and an infected/tumoural cell which must be eliminated [19, 28]. The main interactions between NK cells and target cells are illustrated and commented on in figure 4 and its legend.
KIR genes are located on chromosome 19 and HLA genes on chromosome 6, and therefore the inheritance of these separate families of polymorphic genes generates diverse combinations of pairs of iKIR-HLA class I within individuals [19, 29]. Thus, it is possible that some iKIRs do not find specific ligands. Less than 10% of the population has the four pairs of interacting iKIR-HLA class I described above. Nevertheless, approximately 70% of individuals possess two- to three pairs of iKIR-HLA class I and only 20% have a single pair. However, every NK cell acquires a functional competence, but only if it possesses at least one inhibitory receptor like iKIR which interacts in an adequate way with its ligand [19, 26].
If the cell does not fit this selection criterion, the NK cell stays in a hypoactive state, precluding its activation and defensive action against autologous cells or a potential auto-immune response [26]. Besides, aKIRs seem less essential than iKIRs to the function of NK cells since some individuals do not possess aKIR and are nevertheless healthy [21]. Activating KIR can also bind HLA ligands but less strongly compared to iKIR. Most of the aKIR ligands are still unknown, some aKIR can bind HLA-C as well as inhibitory KIR but with less affinity (like KIR2DS21 with some alleles of the HLA-C2 group and -2DS4 with few alleles mainly from HLA-C1 group) but additional ligands for aKIR are the purpose of intense research [21]. As a general rule, a single NK cell possesses more inhibitory receptors than activating receptors [18, 19].
Role of KIR polymorphism in solid organ transplantation
Alloreactivity of a recipient’s NK cells could participate in the immunological reaction which influences the outcome of solid organ transplantation [29, 30]. NK cells can react against the graft by several mechanisms [29, 30]; 1) If the transplanted cells do not express the same HLA class I molecules as the recipient’s, NK cells detect the “missing-self”, (see fig. 4) become activated and induce lysis of the donor cell; 2) Inflammation caused by surgery during solid organ transplantation induces the expression of stress molecules on graft cells that are recognised by aKIRs (“induced-self killing”) making them susceptible to the NK cell attack [30–32].
A limited number of studies have investigated the influence of KIR, their HLA ligands and the outcome of the transplantation. Some reports have demonstrated a correlation between specific KIR/HLA-C and graft survival in kidney and liver transplantation [9, 33, 34]. However, this effect was not systematically observed [35, 36]. The divergent conclusions could be explained by differences in the set-up of the cohorts and the quality of KIR/HLA typing, and these studies also highlight the complexity of HLA–NK cell interactions as well as the pitfalls of studies based on genetic polymorphisms. A recent report suggests that the role of KIR and their HLA ligand could be important mainly in the context of HLA-A, -B, -DR compatible transplantation [37].
In addition, the MHC genotype of the recipient could also be important for the NK cells to reach functional maturity. This requirement for self-MHC-specific KIR has been termed “licensing” [26]. A patient whose NK cells are licensed for KIR2DL2/3 is not a suitable recipient for an organ from a donor with HLA-C specific for KIR2DL1. The licensing of NK cells depends on KIR/HLA-C specificity, which in turn can vary widely from one population to another [29, 30].
In addition, NK cells could induce an immunological tolerance to the transplanted organ, through the elimination of donor’s APCs, limiting the alloreactive T-cell activation by direct recognition [31, 32, 38]. The exact role of NK cell alloreactivity in solid organ transplantation is still controversial. The mechanisms leading to NK cell activation in the context of solid organ transplantation need to be fully elucidated. The role of NK cells is better known with regard to hematopoietic stem cell transplantation, particularly as far as their implication in grafts against leukaemia (graft-versus-leukaemia [GVL]) is concerned [39, 40].
NK cells can be found in biopsies of renal grafts undergoing acute rejection and more interestingly “NK-type” transcripts are a signature of antibody-mediated-rejection [41]. This observation indirectly implies the participation of NK cells in the immunising response to the graft, especially since they exercise all the necessary functions to activate a rejection.
However, further studies are necessary to assess the clinical importance of NK genetic compatibility as a strategy to improve the survival of renal transplants.
Polymorphism of cytokines
Cytokines are small short-acting proteins that are produced by cells of the immune system. They are essential mediators of the inflammatory and immune responses, and once secreted, they bind to a specific receptor on the surface of the target cell and activate cellular function. They are pleiotropic, often redundant, and they can have a synergic or antagonist action, whether local or systemic [17]. The functions of the cytokines discussed in this review are summarised in table 1. Genetic polymorphisms of cytokine genes may influence their levels of production, their affinity to their specific receptors as well as their activity [42, 43]. Polymorphism may be due to a single nucleotide change at the genetic sequence level (single nucleotide polymorphism, SNP). Such mutations occur in the coding or non-coding regions like the promoter, enhancer, methylation site (epigenetic regulation) or RNA splicing site (fig. 5). Several SNPs have been identified in all cytokine genes [44]. In addition, cytokine receptors are also polymorphic, but this aspect will not be discussed here.

Figure 5
Cytokine polymorphism. Gene encoding a cytokine with representation of two polymorphisms of a single nucleotide (SNPs) in the gene promoter (X) and in the gene itself (X).
The role of specific candidate gene polymorphisms has been investigated in a number of studies, mainly in transplantation between relatives. These studies have yielded controversial results due to small sample sizes (inadequate statistical power), incomplete study designs, and lack of adjustments for multiple comparisons. Specifically in organ transplantation, important variables that are critical for graft survival like HLA compatibility, anti-HLA antibodies (DSA, non-DSA) and degree of immunosuppression, have not been taken into account in studies about cytokine polymorphism and transplantation, and this can explain why most of these studies have given inconclusive results. For example, several studies failed to show significant effects of the genetic polymorphism of cytokines on the survival of renal transplants, particularly for cytokines such as IL-6, IL-10, TNF-α and TGF-β1 [45–50].
As illustrated in table 2, studies on IL-6 -174 G/C – an SNP associated with different IL-6 levels – revealed somewhat disparate results, possibly linked to the pro- and anti-inflammatory properties of this cytokine. Also the variable phenotypes hinder informative comparisons between the different studies. Altogether, it appears that IL-6 SNP genotypes associated with a higher production of IL-6 are more frequently associated with an unfavourable clinical outcome. It is of note, however, that the two “negative” studies are the ones with the largest number of patients.
Among the polymorphisms most frequently targeted for the candidate gene analysis are IL-10-1082, TNFα-308, and TGF-β+10. As illustrated in a previous review [51], the data so far available do not warrant conclusive results, although some trends do emerge.
Several studies have reported a link between IL-10-1082G/A polymorphism and renal transplantation outcome [52], a lower rejection risk being associated with the -1082GG genotype (high IL-10) and a higher rejection incidence conferred by the -1082AA genotype (low IL-10).
Several studies have reported that high TNF-α producer genotypes were associated with higher rates of acute rejection episodes after kidney transplantation [53–55], but this association was not confirmed in later studies [55, 56]. Interestingly, in the largest study published to date, TNF-α-308A (high producer genotype) was associated with lower graft survival rates in re-transplant patients but not in first transplant patients [57]. A single-centre study on 436 patients, in which 9 SNPs were tested at the TNF-α, MCP-1, RANTES, IFN-γ, and TGF-β loci, did not reveal any impact on the outcome of kidney transplantation [56]. In a meta-analysis of 1,087 individual patient data targeting TGF-β, IL-10 and TNFα polymorphisms, only 2 SNP-haplotypes, IL-10-1082/819/-592ACC and TGF-β+10/+25CC, were associated with poor outcome but with OR below 1.5 [46].
The impact of genetic polymorphisms of cytokines on renal transplantation is still strongly debated [43, 58, 59]. When analysing these results, it has to be kept in mind that cytokines are involved in complex inflammatory and immunological cascades and that they form a complicated network. Thus, the effect of an isolated genetic variability in a cytokine may be difficult to pinpoint in view of the complex pathways at play in biological systems such as organ rejection. In solid organ transplantation, the immune response is not only determined by the activities of the recipient’s cytokines but also by those of the donor [45, 60].
As with NK polymorphism, cytokine gene polymorphisms are not a criterion for donor selection or for the identification of patients at higher risk of rejection. Additional studies are necessary to shed more light on the relationship between genetic polymorphism of cytokines and outcome of solid organ transplantation.
|
Table 1: Cytokines and their biological effects. |
|
Cytokines
|
Principal secreted cells
|
Biological and cellular actions
|
| IL-6 |
Macrophages
Endothelial cells
T cells |
Pro-inflammatory and anti-inflammatory properties
Synthesis of proteins of the acute phase by the liver
Proliferation of B cells producing antibodies
Production decrease of IL-1 and TNF-α |
| IL-10 |
Monocytes
T cells (Th2) |
Pro-inflammatory and anti-inflammatory properties
Development inhibition of lymphocytes Th1
Increase of humoural inflammatory response
Production decrease of IL-1, TNF-α and IFN-γ
Anergy of T cells |
| TNF-α |
Macrophages
T cells |
Powerful pro-inflammatory action
Endothelial cells activation and increase of adherence molecules
Increase of vascular permeability
Activation and recruitment of PMN on inflammation site
Fever (hypothalamus)
Synthesis of proteins of the acute phase by the liver
Apoptosis of numerous cell types
Stimulation of angiogenesis
Pro-thrombotic activity |
| TGF-β1 |
|
Stimulation of fibrogenesis
Inhibitors of growth of certain cell types |
| Interleukin-6 = IL-6; Interleukin-10 = IL-10; tumour necrosis factor-alpha = TNF-α; Transforming growth factor-beta 1 = TGF-β1. |
|
Table 2: Associations of patients or donors IL-6 -174G/C SNP genotypes with clinical outcome after kidney transplantation. |
|
Reference
|
Number of patients
|
Genotype
|
Outcome
|
| Kocierz 2011 [58] |
199 |
Patient -174GG/GC |
Increased risk graft loss at 5 y |
| Muller-Steinhardt 2002 [48] |
|
Patient -174GG |
Increased 3 y graft survival |
| Muller-Steinhardt 2004 [61] |
158 |
Patient -597/-572/-194 GGG/GGG
Patient -597/-572/-194 GGG/GCG |
Increased 3 y graft survival
Lower 3 y graft survival |
| Marshall 2001 [60] |
145 |
Donor -174CC (low) |
Increased incidence and severity of acute rejection episodes at 30 days |
| Marshall 2001 [60] |
145 |
Patient -174GG |
No impact on rejection |
| Mittal 2007 [49] |
193 |
Patient -174GG |
Higher risk for end-stage renal disease |
| Nikolova 2008 [43] |
66 |
Donor -174CC (low) |
Association with chronic allograft nephropathy |
| Martin 2009 [50] |
99 |
Patient -174GG/GC |
Increased DSA production |
| Hoffmann 2004 [62] |
242 |
Donor -174GG/GC |
No association with acute rejection |
| Alakulppi 2008 [63] |
772 |
Patient/donor -174GG/GC |
No association with acute rejection, thromboembolism, 1-y graft survival |
Conclusion
It is well established that the most important genetic factor in determining the outcome of the renal transplant is HLA compatibility between donor and recipient, particularly at HLA-A, HLA-B and HLA-DRB1 loci. In addition, the new immunosuppressive drugs do not compensate entirely for the deleterious effect of the HLA incompatibility. On the contrary, the polymorphism of NK cell KIR receptors and cytokine gene polymorphisms may play a role, but its clinical impact is still being debated. Due to the pleiotropic effect of cytokines and of the multigenic origin of post-transplant complications, such polymorphisms should be analysed in combinations. In summary, HLA compatibility is the main criterion in organ allocation programmes and for the time being it is too early to resort to KIR/HLA combinations and cytokine genotypes in the decision algorithm. Furthermore, non-immunological factors such as ischaemia time and duration of dialysis before transplantation have to be taken into account. However, given the limited number of available organs for transplantation, the majority of patients are allocated organs only partially HLA-compatible.
Obviously, additional immunogenetic criteria could make the task even more complex and prolong the time on the waiting list for patients with unfavourable genetic factors. The identification of genetic markers could help to stratify the risk of rejection and lead to administer adapted immunosuppressive treatments. Indeed, it is important to identify recipients at high risk of rejection with the aim of adapting their immunosuppression accordingly, while minimising it for those patients at low risk, in order to avoid the side effects of these drugs and ensure the best possible quality of life for transplanted patients. At present, the recommended strategy to improve the outcome of renal transplants is the association of optimal donor/recipient HLA compatibility and the use of personalised immunosuppressive medication.
Funding / potential competing interests: No financial support and no other potential conflict of interest relevant to this article were reported.
References
1 Clark B, Unsworth DJ. HLA and kidney transplantation. J Clin Pathol. 2010;63:21–5.
2 Aydingoz SE, Takemoto SK, Pinsky BW, Salvalaggio PR, Lentine KL, Willoughby L, et al. The impact of human leukocyte antigen matching on transplant complications and immunosuppression dosage. Hum Immunol. 2007;68:491–9.
3 Wissing KM, Fomegne G, Broeders N, Ghisdal L, Hoang AD, Mikhalski D, et al. HLA mismatches remain risk factors for acute kidney allograft rejection in patients receiving quadruple immunosuppression with anti-interleukin-2 receptor antibodies. Transplantation. 2008;85:411–6.
4 Halloran PF. The clinical importance of alloantibody-mediated rejection. Am J Transplant. 2003;3:639–40.
5 Lefaucheur C, Suberbielle-Boissel C, Hill GS, Nochy D, Andrade J, Antoine C, et al. Clinical relevance of preformed HLA donor-specific antibodies in kidney transplantation. Contrib Nephrol. 2009;162:1–12.
6 Ferrari-Lacraz S, Tiercy JM, Villard J. Detection of anti-HLA antibodies by solid-phase assay in kidney transplantation: friend or foe? Tissue Antigens. 2012;79:315–25.
7 Wujciak T, Opelz G. Evaluation of HLA matching for CREG antigens in Europe. Transplantation. 1999;68:1097–9.
8 Su X, Zenios SA, Chakkera H, Milford EL, Chertow GM. Diminishing significance of HLA matching in kidney transplantation. Am J Transplant. 2004;4:1501–8.
9 Opelz G, Dohler B. Effect of human leukocyte antigen compatibility on kidney graft survival: comparative analysis of two decades. Transplantation. 2007;84:137–43.
10 Meier-Kriesche HU, Scornik JC, Susskind B, Rehman S, Schold JD. A lifetime versus a graft life approach redefines the importance of HLA matching in kidney transplant patients. Transplantation. 2009;88:23–9.
11 Tran TH, Dohler B, Heinold A, Scherer S, Ruhenstroth A, Opelz G. Deleterious impact of mismatching for human leukocyte antigen-C in presensitized recipients of kidney transplants. Transplantation. 2011;92:419–25.
12 Tambur AR, Leventhal JR, Friedewald JJ, Ramon DS. The complexity of human leukocyte antigen (HLA)-DQ antibodies and its effect on virtual crossmatching. Transplantation. 2010;90:1117–24.
13 Kosmoliaptsis V, Sharples LD, Chaudhry A, Johnson RJ, Fuggle SV, Halsall DJ, et al. HLA class I amino acid sequence-based matching after interlocus subtraction and long-term outcome after deceased donor kidney transplantation. Hum Immunol. 2010;71:851–6.
14 Rizzari MD, Suszynski TM, Gillingham KJ, Matas AJ. Consideration of donor age and human leukocyte antigen matching in the setting of multiple potential living kidney donors. Transplantation. 2011;92:70–5.
15 Duquesnoy RJ, Marrari M. HLAMatchmaker-based definition of structural human leukocyte antigen epitopes detected by alloantibodies. Curr Opin Organ Transplant. 2009;14:403–9.
16 Duquesnoy RJ. Antibody-reactive epitope determination with HLAMatchmaker and its clinical applications. Tissue Antigens. 2011;77:525–34.
17 Abbas AK, Lichtman AH. Basic immunology: function and disorders of the immune system. Philadelphia: Saunders; 2001.
18 Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74.
19 Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–14.
20 Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20:311–6.
21 Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics 2007;59:1–15.
22 Moretta L, Biassoni R, Bottino C, Mingari MC, Moretta A. Human NK-cell receptors. Immunol Today. 2000;21:420–2.
23 Uhrberg M. Shaping the human NK cell repertoire: an epigenetic glance at KIR gene regulation. Mol Immunol. 2005;42:471–5.
24 Hadaya K, Avila Y, Valloton L, de Rham C, Bandelier C, Ferrari-Lacraz S, et al. Natural killer cell receptor-repertoire and functions after induction therapy by polyclonal rabbit anti-thymocyte globulin in unsensitized kidney transplant recipients. Clin Immunol. 2010;137:250–60.
25 Moretta L, Bottino C, Pende D, Vitale M, Mingari MC, Moretta A. Human natural killer cells: Molecular mechanisms controlling NK cell activation and tumor cell lysis. Immunol Lett. 2005;100:7–13.
26 Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–10.
27 Moretta L, Bottino C, Pende D, Vitale M, Mingari MC, Moretta A. Human natural killer cells: molecular mechanisms controlling NK cell activation and tumor cell lysis. Immunol Lett. 2005;100:7–13.
28 Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–8.
29 Rajalingam R. Variable interactions of recipient killer cell immunoglobulin-like receptors with self and allogenic human leukocyte antigen class I ligands may influence the outcome of solid organ transplants. Curr Opin Organ Transplant. 2008;13:430–7.
30 Villard J: The role of natural killer cells in human solid organ and tissue transplantation. J Innate Immun. 2011;3:395–402.
31 Van der TW, Bromberg JS. Natural killer cells and the immune response in solid organ transplantation. Am J Transplant. 2010;10:1354–8.
32 Bromberg JS, Heeger PS, Li XC. Evolving paradigms that determine the fate of an allograft. Am J Transplant. 2010;10:1143–8.
33 Kunert K, Seiler M, Mashreghi MF, Klippert K, Schonemann C, Neumann K, et al. KIR/HLA ligand incompatibility in kidney transplantation. Transplantation. 2007;84:1527–33.
34 Hanvesakul R, Spencer N, Cook M, Gunson B, Hathaway M, Brown R, et al. Donor HLA-C genotype has a profound impact on the clinical outcome following liver transplantation. Am J Transplant. 2008;8:1931–41.
35 Tran TH, Mytilineos J, Scherer S, Laux G, Middleton D, Opelz G. Analysis of KIR ligand incompatibility in human renal transplantation. Transplantation. 2005;80:1121–3.
36 Tran TH, Middleton D, Dohler B, Scherer S, Meenagh A, Sleator C, Opelz G. Reassessing the impact of donor HLA-C genotype on long-term liver transplant survival. Am J Transplant. 2009;9:1674–8.
37 van Bergen J, Thompson A, Haasnoot GW, Roodnat JI, de Fijter JW, Claas FH, et al. KIR-ligand mismatches are associated with reduced long-term graft survival in HLA-compatible kidney transplantation. Am J Transplant. 2011;11:1959–64.
38 Yu G, Xu X, Vu MD, Kilpatrick ED, Li XC. NK cells promote transplant tolerance by killing donor antigen-presenting cells. J Exp Med. 2006;203:1851–8.
39 Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333–9.
40 Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–40.
41 Hidalgo LG, Sis B, Sellares J, Campbell PM, Mengel M, Einecke G, et al. NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am J Transplant. 2010;10:1812–22.
42 Kruger B, Schroppel B, Murphy BT. Genetic polymorphisms and the fate of the transplanted organ. Transplant Rev. (Orlando) 2008;22:131–40.
43 Nikolova PN, Ivanova MI, Mihailova SM, Myhailova AP, Baltadjieva DN, Simeonov PL, et al. Cytokine gene polymorphism in kidney transplantation – impact of TGF-beta 1, TNF-alpha and IL-6 on graft outcome. Transpl Immunol. 2008;18:344–8.
44 Tiercy JM. Immunogenetics of hematopoietic stem cell transplantation: the contribution of microsatellite polymorphism studies. Int J Immunogenet. 2011;38:365–72.
45 Marshall SE, McLaren AJ, Haldar NA, Bunce M, Morris PJ, Welsh KI. The impact of recipient cytokine genotype on acute rejection after renal transplantation. Transplantation. 2000;70:1485–91.
46 Thakkinstian A, Dmitrienko S, Gerbase-Delima M, McDaniel DO, Inigo P, Chow KM, et al. Association between cytokine gene polymorphisms and outcomes in renal transplantation: a meta-analysis of individual patient data. Nephrol Dial Transplant. 2008;23:3017–23.
47 Cartwright NH, Keen LJ, Demaine AG, Hurlock NJ, McGonigle RJ, Rowe PA, et al. A study of cytokine gene polymorphisms and protein secretion in renal transplantation. Transpl Immunol. 2001;8:237–44.
48 Muller-Steinhardt M, Hartel C, Muller B, Kirchner H, Fricke L. The interleukin-6 -174promoter polymorphism is associated with long-term kidney allograft survival. Kidney Int. 2002;62:1824–7.
49 Mittal RD, Manchanda PK. Association of interleukin (IL)-4 intron-3 and IL-6 -174 G/C gene polymorphism with susceptibility to end-stage renal disease. Immunogenetics. 2007;59:159–65.
50 Martin J, Worthington J, Harris S, Martin S. The influence of class II transactivator and interleukin-6 polymorphisms on the production of antibodies to donor human leucocyte antigen mismatches in renal allograft recipients. Int J Immunogenet. 2009;36:235–9.
51 Goldfarb-Rumyantzev AS, Naiman N. Genetic prediction of renal transplant outcome. Curr Opin Nephrol Hypertens. 2008;17:573–9.
52 Asderakis A, Sankaran D, Dyer P, Johnson RW, Pravica V, Sinnott PJ, et al. Association of polymorphisms in the human interferon-gamma and interleukin-10 gene with acute and chronic kidney transplant outcome: the cytokine effect on transplantation. Transplantation. 2001;71:674–7.
53 Sankaran D, Asderakis A, Ashraf S, Roberts IS, Short CD, Dyer PA, et al. Cytokine gene polymorphisms predict acute graft rejection following renal transplantation. Kidney Int. 1999;56:281–8.
54 Tinckam K, Rush D, Hutchinson I, Dembinski I, Pravica V, Jeffery J, Nickerson P. The relative importance of cytokine gene polymorphisms in the development of early and late acute rejection and six-month renal allograft pathology. Transplantation. 2005;79:836–41.
55 Alakulppi NS, Kyllonen LE, Jantti VT, Matinlauri IH, Partanen J, Salmela KT, et al. Cytokine gene polymorphisms and risks of acute rejection and delayed graft function after kidney transplantation. Transplantation. 2004;78:1422–8.
56 Brabcova I, Petrasek J, Hribova P, Hyklova K, Bartosova K, Lacha J, Viklicky O. Genetic variability of major inflammatory mediators has no impact on the outcome of kidney transplantation. Transplantation. 2007;84:1037–44.
57 Mytilineos J, Laux G, Opelz G. Relevance of IL10, TGFbeta1, TNFalpha, and IL4Ralpha gene polymorphisms in kidney transplantation: a collaborative transplant study report. Am J Transplant. 2004;4:1684–90.
58 Kocierz M, Siekiera U, Kolonko A, Karkoszka H, Chudek J, Cierpka L, Wiecek A. -174G/C interleukin-6 gene polymorphism and the risk of transplanted kidney failure or graft loss during a 5-year follow-up period. Tissue Antigens. 2011;77:283–90.
59 Dmitrienko S, Hoar DI, Balshaw R, Keown PA. Immune response gene polymorphisms in renal transplant recipients. Transplantation. 2005;80:1773–82.
60 Marshall SE, McLaren AJ, McKinney EF, Bird TG, Haldar NA, Bunce M, et al. Donor cytokine genotype influences the development of acute rejection after renal transplantation. Transplantation. 2001;71:469–76.
61 Muller-Steinhardt M, Fricke L, Muller B, Ebel B, Kirchner H, Hartel C. Cooperative influence of the interleukin-6 promoter polymorphisms -597, -572 and -174 on long-term kidney allograft survival. Am J Transplant. 2004;4:402–6.
62 Hoffmann S, Park J, Jacobson LM, Muehrer RJ, Lorentzen D, Kleiner D, et al. Donor genomics influence graft events: the effect of donor polymorphisms on acute rejection and chronic allograft nephropathy. Kidney Int. 2004;66:1686–93.
63 Alakulppi NS, Kyllonen LE, Partanen J, Salmela KT, Laine JT. Lack of association between thrombosis-associated and cytokine candidate gene polymorphisms and acute rejection or vascular complications after kidney transplantation. Nephrol Dial Transplant. 2008;23:364–8.




