Figure 1
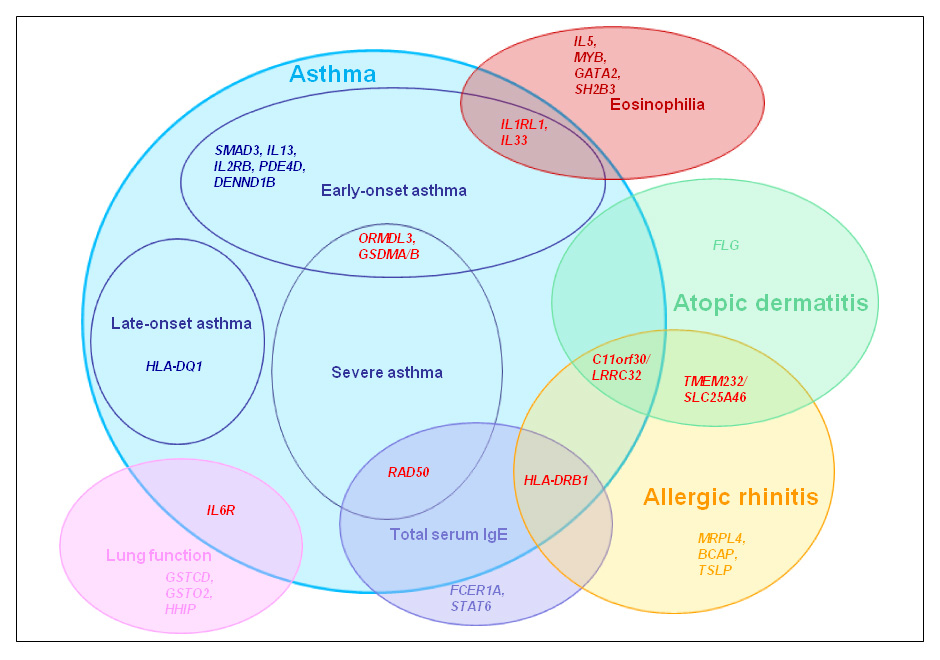
Major susceptibility loci for allergic diseases as identified by GWAS (Venn approach diagram). Genetic loci associated with different subphenotypes are shown in red.
DOI: https://doi.org/10.4414/smw.2012.13612
| Abbreviations | |||
| ACRN | Asthma clinical research network | IL1,2,3,4,5,10,13,33 | Interleukin 1,2,3,4,5,10,13,33 |
| ADRB2 | β2-adrenoreceptor | IL18R1 | Interleukin 18 receptor 1 |
| BCAP | B-cell adaptor for phosphatidylinositol 3-kinase | IL2RB | Interleukin 2 receptor, beta |
| CAMP | Childhood asthma management programme | LABA | Long acting β2-adrenoreceptor agonists |
| CHI3L1 | Chitinase 3-like 1 | LCR | Locus control region |
| CRHR1 | Corticotropin-releasing hormone receptor 1 | MHC | Major histocompatiblity complex |
| DENND1B | DENN/MADD domain containing 1B protein | MRPL4 | Mitochondrial ribosomal protein L4 |
| EDC | Epidermal differentiation complex | NHLBI | National Heart Lung and Blood Institute |
| FCER1A | High affinity receptor for IgE | NK2R | Neurokinin receptor 2 |
| FCER2 | Low affinity receptor for IgE | ORMDL3 | ORM1-like 3 |
| FEF25–75 | Forced expiratory flow 25% and 75% | PDE4D | Phosphodiesterase 4D, cAMP-specific |
| FEV1 | Forced expiratory volume in 1 second | PYHIN1 | Pyrin and HIN domain family member 1 |
| FLG | Fillagrin | RAD50 | DNA repair protein RAD50 |
| FVC | Forced vital capacity | SABAs | Short acting β2-adrenoreceptor agonists |
| GATA2 | GATA binding protein 2 | SH2B3 | SH2B adapter protein 3 |
| GLCCI1 | Glucocorticoid-induced transcript 1 | SNP | Single nucleotide polymorphism |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor | SPT | Skin prick test |
| GSDMA,B | Gasdermin A, B | STAT6 | Signal transducer and activator of transcription 6 |
| GSTO2 | Glutathione S-transferase omega 2 | TBX21 | T box 21 |
| GWAS | Genome-wide Association Studies | TENOR | The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens |
| HHIP | Hedgehog-interacting protein | TSLP | Thymic stromal lymphopoietin |
| HLA | Human leukocyte antigen | V(D)J | Variable, diverse, and joining |
| ICS | Inhaled corticosteroids | ||
Asthma and allergy are complex conditions often present in the same family or closely related subjects. Genetic factors undoubtedly contribute to disease susceptibility but the expression of the disease can be modulated by environmental exposures and the interactions between the two. Candidate-gene and linkage studies followed by positional cloning have already provided a large number of susceptibility genes [1]. The last decade has been marked by the publication of more than 20 genome-wide association studies (GWAS) in asthma or allergy phenotypes. GWAS have reported novel and interesting genes but have also confirmed the role of some functionally relevant genes previously described. However, heritability of allergic diseases has not been elucidated completely so far [2].
The purpose of this review is to evaluate how genetic studies have advanced the understanding of asthma and allergy by identifying specific disease-related markers or sub-phenotypes of the diseases. A search for all published GWAS in allergy-related phenotypes was conducted in MEDLINE using “asthma”, “allergy”, “atopy”, “lung function” and “GWAS” as subject headings. The results were compared with the online catalogue of published GWAS from the National Human Genome Research Institute (http://www.genome.gov/) for the Disease/Trait: “asthma”, “asthma (childhood onset)”, “pulmonary function”, “eosinophil counts”, “IgE levels”, “IgE grass sensitisation”, “YKL-40 levels”, “atopy” and “atopic dermatitis” (version November 2011). In the second part we evaluate how genetic findings may contribute to pharmacogenetics of asthma and allergies.
Asthma is a heterogeneous disease characterised by inflammation of the small airways, bronchial hyper-responsiveness, intermittent airway obstruction, smooth muscle hypertrophy and mucus hypersecretion. Many physiological mechanisms contribute to the disease involving many different cell types [3]. Clinical symptoms differ greatly between patients, suggesting that clinicians are not dealing with a single disease but rather with overlapping conditions of a syndrome [4]. For example atopy, the IgE-mediated response to common allergens in a skin prick test, may be present in many children with asthma but there is a substantial number of patients with non-atopic forms of asthma, where IgE responses to allergens seem not to play a significant role [5].
Descriptive criteria such as age of onset, triggers, frequency and severity of symptoms, as well as response to treatment are often used to classify different sub-phenotypes of asthma. However, these asthma phenotypes are based on the course of the disease and clinical presentation may not reflect distinct disease mechanisms. The discovery of precise markers for specific asthma sub-phenotypes would help greatly in dissecting different entities of the asthma syndrome and would facilitate prevention and treatment of the disease on an individual level.
“Omics” have recently been applied in the quest for such markers in many complex diseases. Genomics is one of the approaches contributing to marker identification. One advantage is that GWAS can systematically interrogate millions of genetic markers across the genome simultaneously and relate them to clinical outcomes or patient characteristics [6] without an a priori hypothesis. Powerful GWAS have now been published involving thousands of subjects and testing association with different disease phenotypes [7].
Childhood onset asthma was investigated in the first published GWAS on asthma in 2007 by the GABRIEL consortium, phase I [8]. A novel locus on chromosome 17q21 was reported to contribute to the susceptibility for early-onset childhood asthma. Associated variants were correlated with mRNA levels of the ORMDL3 gene indicating a genotype-specific regulation of its expression. Other genes on the 17q21 region, namely GSDMA and GSDMB, were later found to also be regulated by the same asthma-associated variants, suggesting that more than one gene from 17q21 may contribute to asthma development [9]. The association of the locus with asthma was robustly replicated in studies involving ethnically diverse populations [10–12].
Chromosomal region 17q21 represents an example of the complexity in delineating the functional variants underpinning the genetic associations. The locus is characterised by high linkage disequilibrium complicating the dissection of the true functional variants from the proxy variants. The co-regulation of ORMDL3 and GSDMA/GSDMB genes by the same variants complicates the identification of the functional gene (or genes) even more. Extensive fine mapping approaches, including complete re-sequencing of the region followed by standardised functional assays for example, are underway to explain the nature of the 17q21 genetic associations [9].
Initial functional studies suggest that ORMDL3 may be implicated in calcium homeostasis which could induce intracellular mechanisms of inflammation [13]. Other studies propose a role of ORMDL proteins in the regulation of sphingolipid metabolism, a hypothesis which needs to be tested further for its relevance in asthma [14]. The observed association between the SNPs at 17q21 locus and other inflammatory diseases such as inflammatory bowel disease and diabetes could also hint to a more general or basic role of this locus in chronic inflammatory conditions [15, 16].
The GABRIEL consortium investigated different forms of asthma, including childhood onset in phase II published in 2010 [17]. In that study, 8,730 asthmatics and 11,389 controls were included; 6,783 were childhood onset asthmatics. The association of the 17q21 locus with childhood onset asthma remained and additional associations with HLA-DQA1, -DQB1, IL33, IL1RL1/IL18R1, SMAD3, IL2RB and IL13were reported, mainly attributable to childhood onset asthma (≤16 years).
In another GWA study, childhood onset asthma was also found to be associated with DENND1B (DENN/MADD domain containing 1B protein) [18]. The study included asthmatics with the severe form of the disease. However, replication of this signal is sparse in other populations which could indicate that it may be related to a specific yet unidentified sub-phenotype of asthma present in the US discovery cohort. The gene is a plausible candidate for a role in asthma pathogenesis as it seems to encode for a protein interacting with TNFα (Tumour necrosis factor α). As DENND1B is expressed by dendritic cells it could very easily play a role in adaptive immune responses [19]. In a second GWAS in a US cohort, the Childhood Asthma Management Program (CAMP) study, a region on chromosome 5q12 was suggested to be associated with childhood asthma, even though the signal did not reach significance in the discovery cohort [20]. Without further fine mapping, the authors concluded that the association signal is related to PDE4D (Phosphodiesterase 4D, cAMP-specific), a gene harboured in the locus. The encoded protein is involved in the airway smooth muscle contractility in knockout mice suggesting that it is a potential therapeutic target [21]. In replication cohorts within the same study the finding remained significant in some study populations of European and Hispanic people, but not in populations of African ancestry.
Childhood asthma was associated with the HLA-DP locus (HLA-DPA1 and HLA-DPB1) in Japanese and Korean populations [22]. In that study, modest associations were shown for the 17q21 locus containing ORMDL3/GSDMB/GSDMA and 5q31 (IL5/RAD50/IL13), whereas there were no associations with PDE4D, DENND1B, IL18R1, and IL2RB.
The GABRIEL consortium GWAS in phase II included 1,947 adult onset asthmatics. HLA-DQ was the only locus presenting strong associations with later onset asthma [17]. A separate analysis of 529 subjects with occupational asthma did not reveal significant results. HLA-DQ/DR variants were also associated with “difficult-to-treat” asthma in TENOR (The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens) GWAS [23]. The study investigated asthmatic adults though it did not provide any information regarding the age of onset of the disease.
GWAS in Asian populations also confirmed genetic heterogeneity between children and adults. The largest GWAS so far published on an Asian population identified the most significant associations between adult asthma and the major histocompatibility complex region (MHC) [24]. The effects were independent of the HLA-DQ associations found in the GABRIEL Consortium GWAS [17].
It is a common belief that the development of IgE-mediated atopy is a precursor for the later development of asthma, a theory also propagated as the atopic march. However, it seems that genetic susceptibility influencing IgE synthesis differs greatly from the susceptibility to developed asthma. Thus, one may speculate that IgE and asthma, while linked to each other, are not linearly related. The idea that asthma just develops on the basis of atopy seems to be too simple in the light of genetic data.
The first GWAS on IgE serum levels identified functional variants on chromosome 1q23 which includes FCER1A (High affinity receptor for IgE) gene, variants within the 5q31 locus, and the 12q13 locus encoding the transcription factor STAT6 [25]. Both FCER1A and STAT6 are functionally involved in IgE regulation, the first being part of the high affinity IgE receptor complex and the second being the intracellular signal from IL4 necessary to induce IgE switching. Both have been extensively studied in atopy and asthma genetics [26, 27]. The 5q31 locus was identified to encompass variants also associated with severe asthma in the TENOR GWAS [23]. The overlapping association of 5q31 locus in IgE [25] and severe asthma [23] could imply that it is involved in atopic asthma. The 5q31 region contains many cytokines genes such as IL3, IL4, IL5, IL13 and GM-CSF, and the RAD50 gene (DNA repair protein RAD50) a less obvious candidate for IgE regulation. The RAD50 gene contains a locus control region (LCR) which could also regulate the transcription of IL4 and IL13 genes [28]. However, RAD50 may itself participate in V(D)J (variable, diverse and joining) and class-switch recombination based on its role in DNA repair and re-ligation [29].
In the GABRIEL consortium study, only HLA-DRB1 was significantly associated with serum IgE levels [17]. These results confirm that genetic determinants of IgE and asthma do not overlap in the populations included in the study. While it is still unclear whether IgE is a secondary event in the development of an asthmatic phenotype, it is obvious that atopic and non-atopic asthma are two diverse entities in terms of genetic susceptibility.
Skin prick test (SPT) and allergen-specific IgE levels have also been analysed for genetic susceptibility signals in a recent GWAS but no consistent patterns of associations were found [30]. In another study performed mainly in adult populations, grass sensitisation was associated with the HLA-DRB4 locus [31]. Taken together, one could hypothesise that genetic determinants for atopy-phenotypes and specific sensitisation differ.
Studying asthma by disease-onset phenotype, severity or atopic status has been the preferred method in GWAS. There are nevertheless studies using intermediate or asthma-related phenotypes in GWAS resulting in statistically strong and plausible outcomes (table 1).
| Table 1: GWAS findings in intermediate asthma phenotypes. | |||
| Phenotype | Genetic locus | Chromosome | Population* |
| YKL-40 levels | CHI3L1 | 1q32 | European [38] |
| Bronchial hyper-responsiveness | |||
| Blood eosinophil counts | SH2B3 | 12q24 | European [34] |
| GATA2 | 3q21 | ||
| IL1RL1 | 2q12 | ||
| IL5 | 5q31 | ||
| IKZF2 | 2q34 | ||
| WDR36 | 5q22 | ||
| MHC | 6p21 | ||
| IL33 | 9p24 | ||
| MYB | 6p22 | ||
| Total serum IgE | FCER1A | 1q23 | European [25] |
| RAD50 | 5q31 | ||
| STAT6 | 12q13 | ||
| Specific IgE levels (HDM, cat fur, mixed grass) | C11orf30/LRRC32 | 11q13 | European [31] |
| FNDC3A | 13q14 | European [30] | |
| Lung function (FEV1 and FVC) | GSTO2 | 10q25 | European [42] |
| Lung function (FEF25–75) | IL6R | 1q21 | |
| Lung function (FEV1 and FEV1/FVC) | HHIP | 4q31 | European [44] |
| GSTCD | 4q24 | ||
| AGER | 6p21 | ||
| THSD4 | 15q23 | ||
| TNS1 | 2q35 | ||
| HTR4 | 5q32 | ||
| DAAM2 | 6p21 | ||
| HHIP | 4q31 | European [45] | |
| AGER/PPT2 | 6p21 | ||
| HTR4 | 5q32 | ||
| ADAM19 | 5q33 | ||
| GPR126 | 6q24 | ||
| FAM13A | 4q22 | ||
| PTCH1 | 9q22 | ||
| * The term populations refers to the ethnicity of the original study. | |||
Eosinophils are important in asthma-related inflammatory responses and high eosinophil counts are a prominent feature of severe asthma [32]. In addition, studies have shown that corticosteroids are more efficient in eosinophilic asthma [33]. An Icelandic study found IL1RL1variants to be associated with higher blood eosinophil counts and to also confer susceptibility to atopic asthma [34]. The replication included nine European populations and one from East Asia. Interestingly, IL1RL1/IL18R1on 2q12 was also associated with childhood asthma in the GABRIEL Consortium [17] and seems to associate with other inflammatory conditions such as Crohn’s disease [35] and atopic dermatitis [36]. This could imply that the IL1 receptor gene cluster on 2q12 also contributes to common mechanisms in inflammatory diseases. In the same GWAS, GATA2 (GATA binding protein 2), IL5 and SH2B3 (SH2B adapter protein 3) were linked to eosinophil counts but not to asthma. All these genes are encoding for proteins expressed in blood cells involved in hematopoietic cell maturation, T cell activation and eosinophil activation, making their role in eosinophilia plausible.
Serum YKL-40 levels may represent a biomarker for asthma and disease severity [37]. YKL-40 serum levels were increased in asthmatics and in those with bronchial hyper-responsiveness compared to controls. A study by Ober et al. used GWA to study genetic involvement in the regulation of YKL-40 levels and found an association between a promoter SNP in CHI3L1 (Chitinase 3-like 1) and YKL-40 levels [38]. CHI3L1 variations were also associated with bronchial hyper-responsiveness and lung function but not atopic state. Despite the compelling role of chitinases in the pathogenesis of asthma [39], CHI3L1 associations with asthma could not yet be replicated in subsequent studies [40].
Reduced pulmonary function is not specific for asthma but plays a role in many obstructive pulmonary diseases. Pulmonary function is a heritable trait depicting the physiological capacity of the lungs [41] and can be viewed as an intermediate and easily measurable phenotype related to asthma. GWAS were conducted in healthy individuals to interrogate the heritability of lung function, and spirometry measurements were tested for genetic associations [42–46]. Variants in the putative genes GSTO2(Glutathione S-transferase omega 2) on chromosome 10 and IL6R (Interleukin 6 receptor) on chromosome 1 showed significant associations with FEV1, FVC and FEF25–75,respectively [42]. Thus, IL6R SNPs are associated with both asthma and lung function [42, 47]. HHIP (Hedgehog-interacting protein) was also identified as a lung function susceptibility locus in a number of well-powered studies [43–46]. HHIP is part of the hedgehog signalling pathway and may play a role in embryonic lung development [48]. However, the reproducible and plausible association of HHIP with lung function was independent of the asthmatic phenotype [43]. Therefore, association of genes with lung function measurements and asthma may identify lung specific mechanisms in asthma development in contrast to general inflammatory mechanisms (such as eosinophilia) that also play a role in asthma development.
Most GWAS have been conducted in populations of European ancestry (tables 1, 2). However, genetic associations with asthma differ significantly between European, African and Asian populations. The 17q21 signal is a good example of this diversity in disease heritability [49, 50]. Association between asthma and 17q21 was well replicated in Europeans and Hispanic populations but not in populations of African ancestry where PYHIN1 (encoding Pyrin and HIN domain family member 1) was identified as a specific risk locus exclusive for this ethnic group. Interestingly, variants in DENND1B showed opposite directions of association with asthma in European and African populations [18]. DENND1B is currently the only asthma candidate gene found to be associated with the disease in both European and African populations.
Together with asthma, atopic dermatitis and allergic rhinitis belong to the group of atopic diseases [51, 52]. These diseases often occur in the same family and thus, a shared genetic susceptibility was suspected for a long time.

Figure 1
Major susceptibility loci for allergic diseases as identified by GWAS (Venn approach diagram). Genetic loci associated with different subphenotypes are shown in red.
Atopic dermatitis often coexists or even precedes asthma. In atopic dermatitis a major gene driving the disease was identified before the GWAS era. The fillagrin (FLG) gene is located in the epidermal differentiation complex (EDC) on chromosome 1q21. It was first associated with ichthyosis vulgaris (common dry skin), before the relationship with atopic eczema was discovered [53]. In cross sectional studies it was shown that FLG accounts for approximately 15% of atopic dermatitis patients in European populations [54]. If and how fillagrin deficiency may influence other atopic phenotypes is still a matter of debate [55].
A GWAS on atopic dermatitis showed significant associations with the 11q35.5 locus (table 2) [56]. The region includes the C11orf30gene which encodes for the nuclear protein EMSY involved in DNA repair [56]. In that GWAS, modest associations were also detected for the EDC, apart from FLG within the region suggesting that other variants in the EDC also contribute to atopic dermatitis. A recent GWAS study in Han Chinese confirmed FLG as a genetic susceptibility locus also in that population. It also uncovered novel susceptibility loci [57]. Ethnic heterogeneity in genetic variants across populations was observed for FLG[58]; however, functional relevant variants in the gene were always associated with atopic dermatitis, independent of ethnicity.
Allergic rhinitis is another expression of an allergic disease with a familial aggregation. A GWAS conducted for allergic rhinitis in a population of Chinese origin showed significant associations with MRPL4 (mitochondrial ribosomal protein L4) on chromosome 19p13.2 and BCAP (B-cell adaptor for phosphatidylinositol 3-kinase) on 10q24.1, in both discovery and replication panels [59]. A suggestive association was detected for the previously atopy-related locus HLA-DQB1/HLA-DRB1[17]. In a meta-analysis of four European populations, variants within the 11q13 locus were associated with allergic rhinitis and grass sensitisation [31]. Harbouring variants in C11orf30 and LRRC32, the locus showed association with atopic dermatitis [56] and atopic asthma [47], respectively. In addition, the previously identified atopic dermatitis susceptibility locus TMEM232/SLC25A46 was also significantly associated with allergic rhinitis [31]. Further suggestive associations were found for SNPs in the TSLP (Thymic stromal lymphopoietin) gene.
Not surprisingly, GWAS reflect the genetic heterogeneity underlying allergic diseases. Although a few shared genetic loci exist among some allergic or asthmatic phenotypes, it is evident that each of these phenotypes has distinctive genetic determinants (fig. 1).
| Table 2: GWAS findings in allergic diseases other than asthma. | |||
| Phenotype | Genetic locus | Chromosome | Population* |
| Atopic dermatitis | C11orf30 | 11q13 | European [56] |
| FLG | 1q21 | European [56], Asian [57] | |
| TMEM232/SLC25A46 | 5q22 | Asian [57] | |
| TNFRSF6B/ZGPAT | 20q13 | ||
| Allergic rhinitis | MRPL4 | 19p13 | Asian [59] |
| BCAP | 10q24 | ||
| HLA-DQB1/HLA-DRB1 | 6p21 | ||
| TMEM232/SLC25A46 | 5q22 | European [31] | |
| TSLP | 13q31 | ||
| C11orf30/LRRC32 | 11q13 | ||
| *The term populations refers to the ethnicity of the original study. | |||
Current asthma treatments are based on inhaled corticosteroids (ICS), long and short acting β2-adrenoreceptor agonists (LABAs and SABAs) as well as leukotriene antagonists. In the vast majority of patients symptoms are well-controlled with these conventional asthma therapies. However, approximately 20% of patients are not responsive to ICS, a phenotype often called “difficult to treat” or severe asthma [60, 61]. It is this form of asthma which shows an increased risk for exacerbations, hospitalisation and death.
Thus, there is a need for markers characterising patients who (1) benefit from a particular treatment and (2) exclude those at risk for side effects. Identification of responders and non-responders to certain (expensive) medications may also reduce costs for the treatment of severe asthma [62]. Pharmacogenetic studies show that genetic variation can determine and modify an individual’s response to drugs. In asthma, most pharmacogenetic studies retrospectively investigated effects of medication on changes in FEV1 or asthma exacerbations in relation to genetic variation in a specific candidate gene (or a candidate pathway), potentially involved in treatment-associated mechanisms. Here, we firstly discuss pharmacogenetic studies related to polymorphisms in the β2-adrenoreceptor (ADRB2) important in SABA and LABA treatment. Then, we describe studies dealing with severe or “difficult to treat” asthma and finally we discuss the potential role of pharmacogenetics in treatment with biologicals, a new group of asthma medication only recently introduced to clinical practice.
Short and long acting β2-adrenoreceptor agonists (SABAs and LABAs) are the most commonly prescribed reliever drugs in asthma, ameliorating bronchoconstriction and leading to long-term symptom control [63]. The primary target of β2-adrenoreceptor agonists is the β2-adrenoreceptor located on the surface of airway smooth muscle cells. The ADRB2gene, coding for this receptor, is a highly polymorphic locus with common and rare SNPs in exonic and regulatory regions of the gene. These SNPs are associated with asthma phenotypes and response to treatment [64].
When clinical and epidemiological studies reported that some patients were experiencing life-threatening symptoms and death following the use of β2-adrenoreceptor agonists [65–67], the need to have a better understanding of individual differences to treatment responses became essential. Three polymorphisms leading to amino acid changes in the receptor, Arg16Gly, Gln27Glu and the rare Thr164Ile, have been the focus of ADRB2 pharmacogenetic studies on SABA and LABA effects.
The Arg16Gly polymorphism is very common in Europeans; approximately 40% of the population carries an Arg16 allele. Patients with the Arg16Arg variation were more responsive to SABA treatment with albuterol [68]. However, Arg16Arg carriers had a reduced response compared to Gly16Gly carriers when albuterol was used regularly [69]. In case of severe asthma exacerbations, children with Gly16Gly showed a better response to albuterol [70]. It was speculated that these differences in response to SABA may be modified by further polymorphisms in the gene such as Gln27Glu [71].
To date it is unclear if the Arg16Gly genotype could be used to predict which patients will respond well to LABA treatment in combination with ICS and which are at increased risk for serious adverse effects. An initial retrospective study by the National Heart Lung and Blood Institute (NHLBI) Asthma Clinical Research Network (ACRN) suggested that patients with Arg16Arg showed a decline in lung function following combined LABA and ICS treatment [72]. The findings of a British cohort study consisting of children under regular use of salmeterol and children not taking salmeterol supported the idea that Arg16Arg carriers have a higher asthma exacerbation risk compared to Gly16Gly carriers [73]. Extending their cohort population, the authors investigated the risk of asthma exacerbations related to Arg16Gly polymorphism in young asthmatics under regular and “on demand” use of albuterol [74]. The increasing exacerbation risk effect of Arg16 was still evident in patients under regular use of SABAs or LABAs. The results emphasise that carriers of Arg16 are at increased risk for exacerbation when either SABAs or LABAs are used as a regular reliever. Later studies consisting of asthmatic adults under combined LABA and ICS treatment showed no pharmacogenetic effect of Arg16Gly [75, 76]. Indeed comparative meta-analysis suggested that children have an augmented risk for adverse effects compared to adults but these differences (potentially due to small sample size in childhood studies) did not reach statistical significance [77].
Gln27Glu is in high linkage disequilibrium with Arg16Gly [78] and thus, Arg16Arg is predominantly combined with Gln27Gln. As a result Gln27Glu could be considered to act as a co-modifier of Arg16Gly effects. The combination of the two polymorphisms seems to affect the binding of the ligand to the ADRB2 receptor and the downstream signal transduction [79–81]. Similarly, Thr164Ile may potentially influence the binding of β2-adrenoreceptor agonists. Despite its low frequency (<3% in European populations), the position of Thr164Ile polymorphism in one of the transmembrane domains suggests that it may affect ADRB2 function [80, 82] in a small fraction of the population carrying the polymorphisms and taking SABA or LABA.
Existing pharmacogenetic studies on ADRB2 have provided initial evidence that genetic factors could be important in patients treated for asthma. However, due to study design, it is difficult to distinguish if ADRB2 polymorphisms increase the risk for exacerbation per se, or if these effects are truly due to pharmacogenetic interaction with SABA and LABA. In prospective pharmacogenetic studies a comparison needs to be made between carriers of different ADRB2 genotypes and use of either β2-adrenoreceptor agonists or anticholinergics.
Inhaled corticosteroids (ICS) represent the main and most effective anti-inflammatory controller treatment in asthma improving bronchial hyper-responsiveness and lung function while reducing asthma exacerbations [63, 83]. Corticosteroid function in asthma is not fully understood. In part, it may act on intracellular glucocorticoid receptors. Corticosteroids can inhibit the expression of pro-inflammatory molecules such as IL5 and IL6 while promoting the expression of regulatory cytokines such as IL10 [84, 85]. Furthermore, corticosteroids are potent regulators of histone acetylation, having significant epigenetic effects [86]. Asthmatic patients do not always respond to ICS and in severe asthma high doses of inhaled or even oral corticosteroids are often administered in an attempt to control symptoms [60, 87].
Three separate candidate gene studies conducted in the CAMP population suggested pharmacogenetic effects of variants in CRHR1 (Corticotropin-releasing hormone receptor 1) [88], TBX21 (T box 21) [89] and FCER2 (Low affinity receptor for IgE) [90] in relation to ICS treatment. CRHR1 variants significantly influenced lung function in patients receiving ICS [88]. However, the result could not be replicated in a following study [91]. In a second study in the same population, a rare SNP leading to an amino acid change in the Th1 cells induction transcription factor T-bet, modulated the effect of ICS treatment on bronchial hyperresponsiveness [89]. In an independent study of mild to moderate Asian asthmatics receiving ICS, asthma control was more easily achieved in patients carrying the wild type alleles of TBX21H33Q and NK2R (Neurokinin receptor 2) G231E polymorphism [92]. As NK2 receptors can mediate bronchoconstriction it was plausible that this genetic variant was associated with increased FEV1% in the same study [92].
Again in the CAMP study, the T2206C polymorphism in the FCER2 gene was associated with severe exacerbations and increased IgE levels in severe asthmatic children receiving ICS but not in those not receiving medication [90]. FCER2 gene encodes for the low affinity receptor for IgE, CD23. CD23 is a major regulator in allergic asthma and could be implicated in persistently elevated IgE levels seen in some asthmatics under ICS [93]. The effect was present in patients from European and African ancestry in the CAMP study. An independent study confirmed that the T2206C variant is a genetic marker for severe exacerbations in children with severe asthma despite increased use of ICS [94].
A recent GWAS aimed to examine pharmacogenetic effects in children on ICS (budesonide) [95]. The study found a number of suggestive hits in a small discovery cohort associated with improvement of lung function measurements after ICS treatment. After replication in additional small patient groups, further investigations focused on one of these polymorphisms located near GLCCI1 (Glucocorticoid-induced transcript 1 gene) [96]. Preliminary molecular studies suggest that polymorphism rs37973 may indeed influence gene function. The authors suggest that this SNP may be a marker for ICS response but further, independent replications and proper functional assessments are necessary before drawing conclusions.
The fact that there are asthma patients who do not respond well to current standard therapy leads to the development of novel drugs addressing specific immune-mechanisms thought to be important in asthma. A biological therapy which has already entered clinical practice is omalizumab, a humanised IgE monoclonal antibody used in both severe to moderate allergic asthma and allergic rhinitis [97, 98]. Treatment with omalizumab resulted in reduction of asthma exacerbations, hospitalisations and use of inhaled corticosteroids as well as improved lung function in a number of studies [99–101]. Serious adverse effects, in particular anaphylactic episodes, have been reported [102]. At present, a series of steps are recommended to be taken by the physicians to avoid anaphylactic reactions [103]. Considering the potential specificity of the action, potential side effects and the considerable cost of treatment, it is surprising how little effort has been made for proper matching of patients to this therapy.
Genetics could be one of the tools used for matching; others will be immunological and clinical profiling. Most likely, a combination of these approaches will be successful [104, 105]. Considering treatment costs of biologicals, matching is necessary and cost effective. It will increase acceptance of these expensive treatments with patients and the public. One obvious factor to be considered should be the T2206C polymorphism in the FCER2 gene [90]. According to recent data, patients with elevated IgE and exacerbations induced by infections may also profit from omalizumab therapy [100].
Other biological therapies are mepolizumab, an anti-IL5 monoclonal antibody; the IL4 variant pitrakinra; and lebrikizumab, a humanised monoclonal antibody against IL13. Clinical trials including patients non-responsive to inhaled or oral corticosteroids and eosinophilic severe asthma showed that anti-IL5 can provide an effective treatment by acting on airway thickening and improving the disease exacerbations [106, 107]. Randomised, double-blind, placebo-controlled clinical trials on anti-IL13 and the IL4 variant resulted in the reduction of therapy-related adverse effects and significant improvement of lung function [108, 109]. In these studies, specific subphenotypes of asthma had been selected for clinical trials based on rather simple selection criteria. It is conceivable that subgroups of patients characterised by an aggravation of genetic variants in the IL4/IL13 pathway would be an even better target population for pitrakinra and lebrikizumab therapies [110].
So far, there are no pharmacogenetic studies published relating to biological treatments. However treatments in severe or “difficult to treat” asthmatics would be facilitated by the identification of molecular markers which could convincingly identify responders and those at increased risk for side effects. In these settings, pharmacogenetic information can be of great clinical importance.
When using genetics it becomes clear that asthma may not be just one disease but many. Different mechanisms may lead to different sub phenotypes, some of which are not sufficiently controlled with current therapy. Thus, genetic studies can contribute to the understanding of the disease and, in a second step, may improve diagnosis and therapy. Modern genetic tools such as GWAS and whole genome sequencing are now feasible in clinical studies and should be applied in pharmacological trials. The ultimate goal would be to apply a personalised approach in respiratory medicine, to which genetics can contribute. This would increase the efficacy and safety of treatment and reduce side effects.
Acknowledgments:The publication was screened for unintentional mistakes in quoting references using Plagiarism Detector software (http://www.plagiarism-detector.com).
1 Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8(3):169–82.
2 Kabesch M. Next generation genetics in allergy. Curr Opin Allergy Clin Immunol. 2010;10(5):407.
3 Bhakta NR, Woodruff PG. Human asthma phenotypes: from the clinic, to cytokines, and back again. Immunol Rev. 2011;242(1):220–32.
4 Kiley J, Smith R, Noel P. Asthma phenotypes. Curr Opin Pulm Med. 2007;13(1):19–23.
5 Bentley AM, Menz G, Storz C, Robinson DS, Bradley B, Jeffery PK, et al. Identification of T lymphocytes, macrophages, and activated eosinophils in the bronchial mucosa in intrinsic asthma. Relationship to symptoms and bronchial responsiveness. Am Rev Respir Dis. 1992;146(2):500–6.
6 Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299(11):1335–44.
7 Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242(1):10–30.
8 Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–3.
9 Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85(3):377–93.
10 Galanter J, Choudhry S, Eng C, Nazario S, Rodriguez-Santana JR, Casal J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177(11):1194–200.
11 Hirota T, Harada M, Sakashita M, Doi S, Miyatake A, Fujita K, et al. Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J Allergy Clin Immunol. 2008;121(3):769–70.
12 Leung TF, Sy HY, Ng MC, Chan IH, Wong GW, Tang NL, et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64(4):621–8.
13 Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19(1):111–21.
14 Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463(7284):1048–53.
15 McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42(4):332–7.
16 Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–7.
17 Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–21.
18 Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362(1):36–44.
19 Del Villar K, Miller CA. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer's disease brain and hippocampal neurons. Proc Natl Acad Sci U S A. 2004;101(12):4210–5.
20 Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84(5):581–93.
21 Mehats C, Jin SL, Wahlstrom J, Law E, Umetsu DT, Conti M. PDE4D plays a critical role in the control of airway smooth muscle contraction. FASEB J. 2003;17(13):1831–41.
22 Noguchi E, Sakamoto H, Hirota T, Ochiai K, Imoto Y, Sakashita M, et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 2011;7(7): e1002170.
23 Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 125(2):328–35 e11.
24 Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43(9):893–6.
25 Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4(8):e1000166.
26 Potaczek DP, Sanak M, Szczeklik A. Additive association between FCER1A and FCER1B genetic polymorphisms and total serum IgE levels. Allergy. 2007;62(9):1095–6.
27 Schedel M, Carr D, Klopp N, Woitsch B, Illig T, Stachel D, et al. A signal transducer and activator of transcription 6 haplotype influences the regulation of serum IgE levels. J Allergy Clin Immunol. 2004;114(5):1100–5.
28 Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19(1):145–53.
29 Potaczek DP, Kabesch M. Current concepts of IgE regulation and impact of genetic determinants. Clin Exp Allergy. 2012 Jan 30.
30 Wan YI, Strachan DP, Evans DM, Henderson J, McKeever T, Holloway JW, et al. A genome-wide association study to identify genetic determinants of atopy in subjects from the United Kingdom. J Allergy Clin Immunol. 2011;127(1):223–31, 31 e1–3.
31 Ramasamy A, Curjuric I, Coin LJ, Kumar A, McArdle WL, Imboden M, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128(5):996–1005.
32 Gibson PG, Saltos N, Borgas T. Airway mast cells and eosinophils correlate with clinical severity and airway hyperresponsiveness in corticosteroid-treated asthma. J Allergy Clin Immunol. 2000;105(4):752–9.
33 Berry M, Morgan A, Shaw DE, Parker D, Green R, Brightling C, et al. Pathological features and inhaled corticosteroid response of eosinophilic and non-eosinophilic asthma. Thorax. 2007;62(12):1043–9.
34 Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41(3):342–7.
35 Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, et al. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82(5):1202–10.
36 Shimizu M, Matsuda A, Yanagisawa K, Hirota T, Akahoshi M, Inomata N, et al. Functional SNPs in the distal promoter of the ST2 gene are associated with atopic dermatitis. Hum Mol Genet. 2005;14(19):2919–27.
37 Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357(20):2016–27.
38 Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358(16):1682–91.
39 Moffatt MF, Cookson WO. Asthma and chitinases. N Engl J Med. 2008;358(16):1725–6.
40 Wu AC, Lasky-Su J, Rogers CA, Klanderman BJ, Litonjua A. Polymorphisms of chitinases are not associated with asthma. J Allergy Clin Immunol. 2010;125(3):754–7, 7 e1–7 e2.
41 Wilk JB, Djousse L, Arnett DK, Rich SS, Province MA, Hunt SC, et al. Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet Epidemiol. 2000;19(1):81–94.
42 Wilk JB, Walter RE, Laramie JM, Gottlieb DJ, O’Connor GT. Framingham Heart Study genome-wide association: results for pulmonary function measures. BMC Med Genet. 2007;8(Suppl 1):S8.
43 Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5(3):e1000429.
44 Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42(1):36–44.
45 Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42(1):45–52.
46 Artigas MS, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011 Sep 25.
47 Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378(9795):1006–14.
48 Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–87.
49 Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125(2):336–46 e4.
50 Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–92.
51 Gustafsson D, Sjoberg O, Foucard T. Development of allergies and asthma in infants and young children with atopic dermatitis – a prospective follow-up to 7 years of age. Allergy. 2000;55(3):240–5.
52 Rhodes HL, Thomas P, Sporik R, Holgate ST, Cogswell JJ. A birth cohort study of subjects at risk of atopy: twenty-two-year follow-up of wheeze and atopic status. Am J Respir Crit Care Med. 2002;165(2):176–80.
53 Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38(4):441–6.
54 Weidinger S, O’Sullivan M, Illig T, Baurecht H, Depner M, Rodriguez E, et al. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol. 2008;121(5):1203–9 e1.
55 Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med. 2011;365(14):1315–27.
56 Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41(5):596–601.
57 Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43(7):690–4.
58 Blumenthal MN, Langefeld CD, Beaty TH, Bleecker ER, Ober C, Lester L, et al. A genome-wide search for allergic response (atopy) genes in three ethnic groups: Collaborative Study on the Genetics of Asthma. Hum Genet. 2004;114(2):157–64.
59 Andiappan AK, Wang de Y, Anantharaman R, Parate PN, Suri BK, Low HQ, et al. Genome-wide association study for atopy and allergic rhinitis in a Singapore Chinese population. PLoS One. 2011 6(5):e19719.
60 Szefler SJ, Martin RJ, King TS, Boushey HA, Cherniack RM, Chinchilli VM, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002;109(3):410–8.
61 Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005;115(2):233–42.
62 Barnett SB, Nurmagambetov TA. Costs of asthma in the United States: 2002–2007. J Allergy Clin Immunol. 2011;127(1):145–52.
63 Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 2007;120(5 Suppl):S94–138.
64 Litonjua AA. The significance of beta2-adrenergic receptor polymorphisms in asthma. Curr Opin Pulm Med. 2006;12(1):12–7.
65 Stolley PD. Asthma mortality. Why the United States was spared an epidemic of deaths due to asthma. Am Rev Respir Dis. 1972;105(6):883–90.
66 Nelson HS, Weiss ST, Bleecker ER, Yancey SW, Dorinsky PM. The Salmeterol Multicenter Asthma Research Trial: a comparison of usual pharmacotherapy for asthma or usual pharmacotherapy plus salmeterol. Chest. 2006;129(1):15–26.
67 Martinez FD. Safety of long-acting beta-agonists – an urgent need to clear the air. N Engl J Med. 2005;353(25):2637–9.
68 Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100(12):3184–8.
69 Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, et al. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162(1):75–80.
70 Carroll CL, Stoltz P, Schramm CM, Zucker AR. Beta2-adrenergic receptor polymorphisms affect response to treatment in children with severe asthma exacerbations. Chest. 2009;135(5):1186–92.
71 Martin AC, Zhang G, Rueter K, Khoo SK, Bizzintino J, Hayden CM, et al. Beta2-adrenoceptor polymorphisms predict response to beta2-agonists in children with acute asthma. J Asthma. 2008;45(5):383–8.
72 Wechsler ME, Lehman E, Lazarus SC, Lemanske RF, Jr., Boushey HA, Deykin A, et al. beta-Adrenergic receptor polymorphisms and response to salmeterol. Am J Respir Crit Care Med. 2006;173(5):519–26.
73 Palmer CN, Lipworth BJ, Lee S, Ismail T, Macgregor DF, Mukhopadhyay S. Arginine-16 beta2 adrenoceptor genotype predisposes to exacerbations in young asthmatics taking regular salmeterol. Thorax. 2006;61(11):940–4.
74 Basu K, Palmer CN, Tavendale R, Lipworth BJ, Mukhopadhyay S. Adrenergic beta(2)-receptor genotype predisposes to exacerbations in steroid-treated asthmatic patients taking frequent albuterol or salmeterol. J Allergy Clin Immunol. 2009;124(6):1188–94 e3.
75 Bleecker ER, Yancey SW, Baitinger LA, Edwards LD, Klotsman M, Anderson WH, et al. Salmeterol response is not affected by beta2-adrenergic receptor genotype in subjects with persistent asthma. J Allergy Clin Immunol. 2006;118(4):809-16.
76 Bleecker ER, Postma DS, Lawrance RM, Meyers DA, Ambrose HJ, Goldman M. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007;370(9605):2118–25.
77 Kramer JM. Balancing the benefits and risks of inhaled long-acting beta-agonists – the influence of values. N Engl J Med. 2009;360(16):1592–5.
78 Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000;97(19):10483–8.
79 Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33(32):9414–9.
80 Green SA, Cole G, Jacinto M, Innis M, Liggett SB. A polymorphism of the human beta 2-adrenergic receptor within the fourth transmembrane domain alters ligand binding and functional properties of the receptor. J Biol Chem. 1993;268(31):23116–21.
81 Moore PE, Laporte JD, Abraham JH, Schwartzman IN, Yandava CN, Silverman ES, et al. Polymorphism of the beta(2)-adrenergic receptor gene and desensitization in human airway smooth muscle. Am J Respir Crit Care Med. 2000;162(6):2117–24.
82 Green SA, Rathz DA, Schuster AJ, Liggett SB. The Ile164 beta(2)-adrenoceptor polymorphism alters salmeterol exosite binding and conventional agonist coupling to G(s). Eur J Pharmacol. 2001;421(3):141–7.
83 Barnes PJ, Pedersen S, Busse WW. Efficacy and safety of inhaled corticosteroids. New developments. Am J Respir Crit Care Med. 1998;157(3 Pt 2):S1–53.
84 Robinson D, Hamid Q, Ying S, Bentley A, Assoufi B, Durham S, et al. Prednisolone treatment in asthma is associated with modulation of bronchoalveolar lavage cell interleukin-4, interleukin-5, and interferon-gamma cytokine gene expression. Am Rev Respir Dis. 1993;148(2):401–6.
85 John M, Lim S, Seybold J, Jose P, Robichaud A, O’Connor B, et al. Inhaled corticosteroids increase interleukin-10 but reduce macrophage inflammatory protein-1alpha, granulocyte-macrophage colony-stimulating factor, and interferon-gamma release from alveolar macrophages in asthma. Am J Respir Crit Care Med. 1998;157(1):256–62.
86 Adcock IM, Ford P, Ito K, Barnes PJ. Epigenetics and airways disease. Respir Res. 2006;7:21.
87 Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56(4):1054–70.
88 Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13(13):1353–9.
89 Tantisira KG, Hwang ES, Raby BA, Silverman ES, Lake SL, Richter BG, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A. 2004;101(52):18099–104.
90 Tantisira KG, Silverman ES, Mariani TJ, Xu J, Richter BG, Klanderman BJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007;120(6):1285–91.
91 Dijkstra A, Koppelman GH, Vonk JM, Bruinenberg M, Schouten JP, Postma DS. Pharmacogenomics and outcome of asthma: no clinical application for long-term steroid effects by CRHR1 polymorphisms. J Allergy Clin Immunol. 2008;121(6):1510–3.
92 Ye YM, Lee HY, Kim SH, Jee YK, Lee SK, Lee SH, et al. Pharmacogenetic study of the effects of NK2R G231E G>A and TBX21 H33Q C>G polymorphisms on asthma control with inhaled corticosteroid treatment. J Clin Pharm Ther. 2009;34(6):693–701.
93 Wu CY, Sarfati M, Heusser C, Fournier S, Rubio-Trujillo M, Peleman R, et al. Glucocorticoids increase the synthesis of immunoglobulin E by interleukin 4-stimulated human lymphocytes. J Clin Invest. 1991;87(3):870–7.
94 Koster ES, Maitland-van der Zee AH, Tavendale R, Mukhopadhyay S, Vijverberg SJ, Raaijmakers JA, et al. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy. 2011;66(12):1546–52.
95 Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365(13):1173–83.
96 Chapman MS, Askew DJ, Kuscuoglu U, Miesfeld RL. Transcriptional control of steroid-regulated apoptosis in murine thymoma cells. Mol Endocrinol. 1996;10(8):967–78.
97 Hanania NA, Alpan O, Hamilos DL, Condemi JJ, Reyes-Rivera I, Zhu J, et al. Omalizumab in severe allergic asthma inadequately controlled with standard therapy: a randomized trial. Ann Intern Med. 2011;154(9):573–82.
98 Casale TB, Stokes JR. Future forms of immunotherapy. J Allergy Clin Immunol. 2011;127(1):8–15; quiz 6–7.
99 Walker S, Monteil M, Phelan K, Lasserson TJ, Walters EH. Anti-IgE for chronic asthma in adults and children. Cochrane Database Syst Rev. 2006 (2):CD003559.
100 Busse WW, Morgan WJ, Gergen PJ, Mitchell HE, Gern JE, Liu AH, et al. Randomized trial of omalizumab (anti-IgE) for asthma in inner-city children. N Engl J Med. 2011;364(11):1005–15.
101 Rodrigo GJ, Neffen H, Castro-Rodriguez JA. Efficacy and safety of subcutaneous omalizumab vs placebo as add-on therapy to corticosteroids for children and adults with asthma: a systematic review. Chest. 2011;139(1):28–35.
102 Lin RY, Rodriguez-Baez G, Bhargave GA. Omalizumab-associated anaphylactic reactions reported between January 2007 and June 2008. Ann Allergy Asthma Immunol. 2009;103(5):442–5.
103 Cox L, Lieberman P, Wallace D, Simons FE, Finegold I, Platts-Mills T, et al. American Academy of Allergy, Asthma & Immunology/American College of Allergy, Asthma & Immunology Omalizumab-Associated Anaphylaxis Joint Task Force follow-up report. J Allergy Clin Immunol. 2011;128(1):210–2.
104 Knuffman JE, Sorkness CA, Lemanske RF, Jr., Mauger DT, Boehmer SJ, Martinez FD, et al. Phenotypic predictors of long-term response to inhaled corticosteroid and leukotriene modifier therapies in pediatric asthma. J Allergy Clin Immunol. 2009;123(2):411–6.
105 Tantisira KG, Drazen JM. Genetics and pharmacogenetics of the leukotriene pathway. J Allergy Clin Immunol. 2009;124(3):422–7.
106 Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360(10):973–84.
107 Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, et al. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360(10):985–93.
108 Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365(12):1088–98.
109 Wenzel S, Wilbraham D, Fuller R, Getz EB, Longphre M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet. 2007;370(9596):1422–31.
110 Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, et al. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117(2):269–74.
Funding / potential competing interests: This work was funded by the German Ministry of Education and Research (BMBF) as part of the National Genome Research Network (NGFN), with grant NGFN 01GS0810 and the German Research Council (DFG) by grant SFB587 project B16. All authors declare that they have no competing financial or personal interests.