Figure 1
DOI: https://doi.org/10.4414/smw.2012.13521
Parkinson’s disease (PD) is the second most common neurodegenerative disease. It is characterised by the selective loss of dopamine neurons in the substantia nigra compacta. The relative role of the pathogenic gene contributing to the risk of PD has been under considerable debate. Paired-like homeodomain transcription factor 3 (PITX3) is expressed during development in mesencephalic dopaminergic neurons in both mice and humans [1] and it is maintained only in those midbrain neurons that contain dopamine [2]. PITX3 has been reported to be involved in the differentiation of midbrain dopaminergic neurons [3]. When it is disrupted, the dopaminergic neurons in the substantia nigra are selectively lost [4]. Recently SNP (rs3758549) in the putative promoter region of the PITX3 gene was identified as being strong associated with PD [5]. Two other PITX3 SNPs (rs2281983 and rs4919621) were reported to be significantly higher in sporadic PD, which resulted in substitutions of C>T in rs2281983 and A>T in rs4919621 [6–7].
In this study, we analysed these three previous PITX3 SNPs (rs2281983, rs4919621 and rs3758549) in 356 PD patients. We found those SNPs associated with PD could not be replicated in Chinese PD patients. Furthermore, we also investigated the association of novel polymorphisms in PITX3 gene with PD. Our data show a strong association of a novel polymorphism c.219G>A (p = 0.000307, OR = 4.48) in the PITX3 Exon 3 with PD in Chinese Han populations.
A total of 656 study subjects were included in this study, comprising 356 patients with PD (221 males and 135 females) and 300 control subjects with no evidence of PD (189 males and 111 females). All patients were recruited from the Department of Neurology at Sir Run Run Shaw hospital affiliated with Zhejiang University School of Medicine and received a standard neurological examination. The PD cases had a mean age of 68.56 ± 9.74 years (age at onset 58.74 ± 10.13). The clinical diagnosis of PD was confirmed by the senior neurologist specialising in movement disorders according to the Consensus Statement of the Movement Disorders Society in 1998 [8]. The control subjects (mean age of 67.79 ± 10.48 years old) were free of neurological disorders. Gender and age proportion between cases and controls was statistically matched. All subjects were of Chinese Han ethnic background. Informed consent for participation in the study was obtained either directly or from his or her guardian in all subjects and the work received approval from the ethics committee of the institution.
To find common SNPs, specific primers for the PITX3 sequences (NM_005029) were designed to screen the exons and the flanking regions including the promoter region (1500 bp upstream from start codon) of PITX3. Information on the primer sequences is provided in table 1. The screening panel included 16 affected PD patients and 10 unaffected individuals. PCR conditions were set as follows: 40 cycles of 30 s denaturation at 95℃, 30 s annealing at 55℃, 60 s extension at 72℃. The amplification products were sequenced using the ABI-3100 platform (Applied Biosystems, Foster City, USA). The results were analysed using the ABI Prism GeneScan and Genotyper program.
Genetic association and the Hardy-Weinberg equilibrium for the distribution of genotypes were tested by χ2 analysis with Yates’ correction. Odds ratios (ORs) were calculated with 95% confidence intervals (CI). The pairwise linkage disequilibrium (LD) coefficient r2 was calculated using the programme Haploview (http://www.broad.mit.edu/mpg/haploview/index.php). We defined pairs to be in strong LD if one side upper 95% confidence bound on D’ is >0.98 and lower bound is >0.7. Fraction of strong LD in informative comparisons must be at least 0.95.
| Table 1: Overview of the 13 SNPs of the human PITX3gene that were genotyped in our study by direct sequencing. | ||||
| SNP Namea | Location | Polymorphism | AA Variation | dbSNP IDb |
| c.286C>T | Exon 3 | C/T | – | rs2281983 |
| Intron 1 | A/T | – | rs4919621 | |
| promoter | C/T | – | rs3758549 | |
| c.219G>A | Exon 3 | G/A | – | Novel |
| Intron 1 | A/T | – | rs733283 | |
| Intron 1 | C/G | – | rs3808938 | |
| Intron 1 | A/G | – | rs3758553 | |
| Intron 1 | A/G | – | rs114952263 | |
| c.38G>A | Exon 2 | G/A | Ser/Asn | rs104894175 |
| Intron 1 | C/G | – | rs77381320 | |
| Intron 1 | A/G | – | rs77276404 | |
| Intron 1 | C/T | – | rs75646556 | |
| Intron 1 | A/G | – | rs75395522 | |
| Intron 1 | C/G | – | rs75337770 | |
| Notes. a. Offsets for each SNP were calculated relative to the translation start site of the gene PITX3 mRNA according to GenBank accession number NM_005029. Mutation nomenclature was verified using the Mutalyzer programme (www.lovd.nl/mutalyzer/1.0.1/ http://www.lovd.nl/mutalyzer/ ). b. The ID was taken directly from the public SNP database ( http://www.ncbi.nlm.nih.gov/SNP ). | ||||
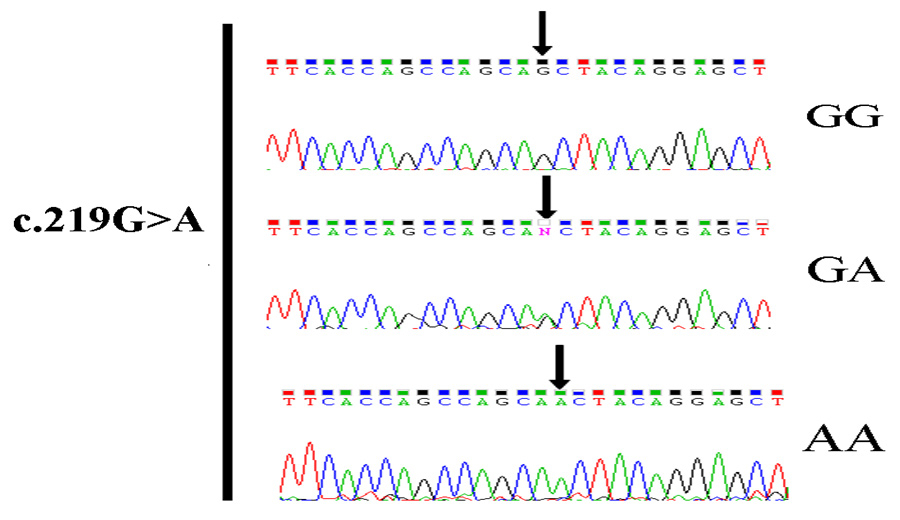
The initial sequence analysis of the promoter region, the introns, and the exons of the PITX3 gene in 16 PD patients revealed no mutations and all polymorphisms except c.219G>A were included in the HapMap data base (NCBI dbSNP build 132) (table 1) (S fig. 1). The finding of a relatively large number of subjects carrying the uncommon variant of the rs2281983 and c.219G>A in PITX3 prompted us to conduct an association study concerning these SNPs in 356 PD patients and 300 healthy controls. Moreover, a HapMap tagging SNP rs4919621 in intron 1 of PITX3 and an SNP rs3758549 in the putative promoter region of the PITX3 gene were also assessed. The genotype distributions of all three investigated polymorphisms were in Hardy-Weinberg equilibrium. The allele distribution is also displayed. When PD patients were compared with controls, no significant differences in genotype or allele frequencies were observed for any of the investigated SNPs (rs2281983, rs4919621 and rs3758549). However, a novel SNP G>A of the c.219 in PITX3 Exon 3 was significantly more common in PD patients than in controls (table 2). In line with these results, a Kaplan-Meier analysis comprising all PD patients confirmed that the c.219 AA genotype of PITX3 is associated with PD more than the other genotypes (p = 0.000307, OR = 4.48). Those patients carrying c.219G>A are all without a family history of PD and with a mean onset age at 54.45 ± 5.98 years. Two patients had tremor as the first symptom and in the other three patients bradykinesia appeared at onset.

Figure 1
| Table 2: Analyses of association of PITX3 Polymorphisms with PD. | ||||||||||
| SNP1 | Location | n2 | Genotype frequency | Allele frequency | ||||||
| Genotype | P-value3 | OR (95%CI)4 | Allele | P-value5 | OR (95%CI)4 | |||||
| PITX3 New Report C.219G>A | Exon3 | GG | G/A | AA | 0.00037 | 4.48 (1.75–11.45) | A | 0.000227 | 4.54 (1.80–11.4) | |
| Cases | 356 | 0.830 | 0.156 | 0.014 | 0.093 | |||||
| Controls | 300 | 0.956 | 0.044 | 0.000 | 0.021 | |||||
| PITX3 SNP 2281983 | Exon3 | CC | C/T | TT | 0.233 | 0.69 (0.39–1.22) | T | 0.274 | 1.26 (0.85–1.87) | |
| Cases | 356 | 0.311 | 0.497 | 0.192 | 0.441 | |||||
| Controls | 300 | 0.393 | 0.443 | 0.164 | 0.385 | |||||
| PITX3 SNP Rs3758549 | Promoter | TT | T/C | CC | 0.890 | 1.05 (0.61–1.82) | C | 0.621 | 0.90 (0.61–1.32) | |
| Cases | 356 | 0.403 | 0.432 | 0.165 | 0.381 | |||||
| Controls | 300 | 0.391 | 0.406 | 0.203 | 0.406 | |||||
| PITX3 SNP Rs4919621 | Intron | AA | A/T | TT | 0.233 | 0.69 (0.39–1.22) | T | 0.274 | 1.26 (0.85–1.87) | |
| Cases | 356 | 0.311 | 0.497 | 0.192 | 0.441 | |||||
| Controls | 300 | 0.393 | 0.443 | 0.164 | 0.385 | |||||
| Notes. 1 SNP name for genotype in cases and controls. 2Number of valid subjects who were successfully genotyped for each of SNP. 3Analysis performed by a 2 x 2 table for each SNP using major homozygotes vs. others in cases and controls. 4Reference group (controls) designated with an OR of 1.00. 5Analysis performed by a 2 x 2 table for the number of each allele in cases and controls. | ||||||||||
Several transcription factors of the homeodomain family critically contribute to the development and the survival of mDA neurons [5, 9]. This led researchers to hypothesise that genetic variants in and around these genes including Engrailed 1/2 [9], PITX3 [5], LMX1B [10] and OTX2 [11] might influence the risk to develop PD. Recently, the association of PITX3 polymorphisms with PD was described in a different patient sample. Fuchs et al provided evidence for the strong association of the PITX3 promoter SNP rs3758549 with PD [5]. Bergman et al. suggested the A-allele of a HapMap tagging SNP rs4919621 was significantly more common in PD patients with an early age of onset, whilst the association of rs3758549 with PD could not be replicated [12]. Haubenberger et al found a strong association between the PITX3 promoter rs3758549 polymorphism and PD (p = 0.0001), as well as an association between EN1 rs1438852 and PD [13]. Le et al showed that PITX3 SNPs rs2281983 and rs4919621 are significantly higher in PD [6].
Here, in our Chinese population of PD patients, we investigated three previously found PITX3 SNPs (rs2281983, rs4919621 and rs3758549) in 356 PD patients. No significant association of these SNPs with PD could be replicated in Chinese Han PD patients. In addition, we also investigated the association of a novel polymorphism c.219G>A in the PITX3 gene with PD. Our data showed a strong association of this polymorphism c.219G>A (p = 0.000307) in the PITX3 Exon 3 with PD in Chinese populations. Although the c.219G>A polymorphism could not influence any amino acids substitution, our case-control association assay showed it could explain a 4.48 increase in risk susceptible to PD pathogenesis when people carry an A type allele. As far as we know, no SNP causing an amino acid shift has been reported in the PITX3 gene. Since the SNPs included in this association study do not result in a change in amino acid sequence, it may be questioned whether they per se influence the function of the protein. However, synonymous SNPs can have consequences for transcription factor binding sites and native splicing sites [14]. Also, a synonymous SNP leading to an infrequent codon in the mRNA sequence appears to be able to affect translation kinetics, causing ribosomes to produce a protein with an altered conformation [15].
Recently, Yu et al. and Liu et al. reported that the PITX3 gene rs3758549 polymorphism may increase the susceptibility of PD in a Chinese population [16, 17]. However, this relationship was not reproduced by a new published study [18], similar to our result. One could speculate on population-specific differences of the general genetic background or environmental factors. Another explanation for this discrepancy could derive from a possibility that an overdominant effect model could explain the results similar to the recessive risk-allele model [5, 16]. Alternative genetic mechanisms such as overdominance might reconcile the seemingly contradictory findings of recessive models.
Furthermore, different diet-lifestyle and regional use of intoxicant chemicals may also play important roles in the risk of PD.
To summarise, our study suggests that novel polymorphisms in the PITX3 gene may be associated with PD in Chinese populations. However, in what particular way this described genetic variants impose their effect in the presymtomatic and symptomatic phase of PD remains to be investigated. Nevertheless, PITX3 may be of importance for PD well in line with recent preclinical and clinical data.
Acknowledgements:The authors thank all the patients who participated in this study and the staff of the Department of Neurology, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, for technical support during the study.
1 Smidt MP, Smits SM, Bouwmeester H, Hamers FP, van der Linden AJ, Hellemons AJ, et al. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–55.
2 Nunes I, Tovmasian LT, Silva RM, Burke RE, Goff SP. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci. U S A 2003;100:4245–50.
3 Smits SM, Smidt MP. The role of Pitx3 in survival of midbrain dopaminergic neurons. J Neural Transm Suppl. 2006:57–60.
4 Hwang DY, Ardayfio P, Kang UJ, Semina EV, Kim KS. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Brain Res Mol Brain Res. 2003;114:123–31.
5 Fuchs J, Mueller JC, Lichtner P, Schulte C, Munz M, Berg D, et al. The transcription factor PITX3 is associated with sporadic Parkinson’s disease. Neurobiol Aging. 2007;30:731–8.
6 Le W, Nguyen D, Lin XW, Rawal P, Huang M, Ding Y, et al. Transcription factor PITX3 gene in Parkinson’s disease. Neurobiol Aging. 2011;32:750–3.
7 Li J, Dani JA, Le W. The role of transcription factor Pitx3 in dopamine neuron development and Parkinson’s disease. Curr Top Med Chem. 2009;9:855–9.
8 Deuschl G, Bain P, Brin M. Consensus statement of the Movement Disorder Society on Tremor. Ad Hoc Scientific Committee. Mov Disord. 1998;13(Suppl 3):2–23.
9 Le Pen G, Sonnier L, Hartmann A, Bizot JC, Trovero F, Krebs MO, et al. Progressive loss of dopaminergic neurons in the ventral midbrain of adult mice heterozygote for Engrailed1: a new genetic model for Parkinson’s disease? Parkinsonism Relat Disord. 2008;14(Suppl 2):S107–111.
10 Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–41.
11 Schilter KF, Schneider A, Bardakjian T, Soucy JF, Tyler RC, Reis LM, et al. OTX2 microphthalmia syndrome: four novel mutations and delineation of a phenotype. Clin Genet. 2011;79(2):158–68.
12 Bergman O, Hakansson A, Westberg L, Nordenström K, Carmine Belin A, Sydow O, et al. PITX3 polymorphism is associated with early onset Parkinson’s disease. Neurobiol Aging. 2010; 31:114–7.
13 Haubenberger D, Reinthaler E, Mueller JC, Pirker W, Katzenschlager R, Froehlich R, et al. Association of transcription factor polymorphisms PITX3 and EN1 with Parkinson’s disease. Neurobiol Aging. 2011;32:302–7.
14 Thi Tran HT, Takeshima Y, Surono A, Yagi M, Wada H, Matsuo M. A G-to-A transition at the fifth position of intron-32 of the dystrophin gene inactivates a splice-donor site both in vivo and in vitro. Mol Genet Metab. 2005;85:213–9.
15 Komar AA. Genetics. SNPs, silent but not invisible. Science. 2007;315:466–7.
16 Yu LH, Lin ZF, Liu Y, Hu FY, He XH, Liu ZL, et al. The transcription factor Pitx3 is a risk modifier for Parkinson’s disease in a Chinese Han population. Eur J Neurol. 2011;18(5):778–83.
17 Liu J, Sun QY, Tang BS, Hu L, Yu RH, Wang L, et al. PITX3 gene polymorphism is associated with Parkinson’s disease in Chinese population. Brain Res. 2011;1392:116–20.
18 Cai Y, Ding H, Gu Z, Ma J, Chan P. Genetic variants of the PITX3 gene are not associated with late-onset sporadic Parkinson’s disease in a Chinese population. Neurosci Lett. 2011;498(2):124–6.
Funding / potential competing interests: No financial support and no other potential conflict of interest relevant to this article were reported.