Genetic polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer (NSCLC) patients
DOI: https://doi.org/10.4414/smw.2011.13275
F
zhou, Z
yu, T
jiang, H
lv, R
yao, J
liang
Summary
PRINCIPLES: Platinum agents cause DNA cross-linking and oxidative damage. Genetic polymorphisms of GSTP1 and XRCC1 involved in glutathione metabolic and DNA repair pathways may explain inter individual differences in chemosensitivity and clinical outcome in NSCLC patients treated with platinum-based regimens.
METHODS: We used DNA sequencing methods to evaluate genetic polymorphisms of the GSTP1A313G and XRCC1G28152A in 111 patients with stage IV NSCLC treated with platinum-based chemotherapy. Clinical response was evaluated according to RECIST criteria after 2–3 cycles of chemotherapy and time to progression (TTP) was calculated from the time of initial treatment to disease progression.
RESULTS: GSTP1A313G and XRCC1G28152A polymorphisms, both alone and in combination, were significantly associated with response to treatment and clinical outcome (p<0.05) in NSCLC patients treated with platinum-based chemotherapy. These polymorphisms independently predicted clinical outcome even after taking into account age, gender, tumour histology, tumour differentiation and chemotherapy regimens.
CONCLUSION: Genetic polymorphisms of GSTP1 and XRCC1 may be important predictive factors in platinum-treated patients with advanced NSCLC. Assessment of genetic variations of GSTP1 and XRCC1 could facilitate therapeutic decisions for individualised therapy in advanced NSCLC.
Introduction
Lung cancer is one of the most prevalent cancers worldwide, and it has become the leading death cause in cities in China [1]. About 80% of lung cancer patients are diagnosed as non-small cell lung cancer (NSCLC), of whom approximately two thirds are diagnosed in advanced stages [2]. Chemotherapy has been the mainstay of treatment for advanced NSCLC. Platinum-based doublets are used as standard first line chemotherapy in NSCLC patients, with an objective response rate of about 40%, a median survival time of 8–10 months and a one-year survival rate of 30–40% [3, 4]. However, even patients with the same demographic and clinical characteristics usually display different responses and varied prognosis. Thus, the concept of individualised chemotherapy based on pharmacogenetics and pharmacogenomics becomes more intriguing, and one of the remaining challenges is the development of predictive markers to individualise and optimise patient therapies.
Platinum agents remain the most commonly used chemotherapeutic agents in advanced NSCLC patients and are known to act through the formation of bulky intra- and interstrand DNA adducts that inhibit DNA synthesis and transcription. Proposed mechanisms of resistance to platinum agents have been attributed to inactivation of platinum compounds through the glutathione metabolic pathway and increased tolerance to DNA damage as a consequence of enhanced DNA repair capacity. It is more likely that it is the combination of these mechanisms that results in cisplatin resistance. Glutathione S-transferases (GSTs) are phase II metabolic enzymes that are involved in the detoxification of mutagenic and cytotoxic DNA-reactive molecules mediated by glutathione conjugation. Currently, many of the commonly used drugs in lung cancer chemotherapy are metabolised by the glutathione system, especially the platinum drugs [5]. Glutathione S-transferase P1 (GSTP1) is a subclass of GSTs that directly participates in the detoxification of platinum compounds and is an important mediator of both intrinsic and acquired resistance to platinum [6]. A single nucleotide substitution (A→G) at position 313, which results in replacement of isoleucine (Ile) with valine (Val) at codon 105, has been found to modify enzyme activity and affinity for electrophilic substrates [7]. Previous studies have shown that expression level and the polymorphic A313G of GSTP1 are linked to the sensitivity of cancer cells to platinum [8]. It has also been demonstrated that enhanced DNA repair capacity is further critical mechanism of resistance to platinum-based chemotherapy that leads to the removal of cisplatin-DNA adducts. Genetic polymorphisms of DNA repair genes are therefore obviously good candidates for determinants of repair capacity and chemotherapy efficacy. X-ray cross-complementing group 1 (XRCC1) is a key enzyme in the gap-filling step of the short patch base excision repair (BER) pathway. The BER pathway, which is involved in protecting cells from the deleterious effects of endogenous DNA damage induced by hydrolysis, reactive oxygen species and other intracellular metabolites that modify DNA base structure, is also important in resisting lesions produced by ionizing radiation and strong alkylating agents, such as platinum agents. The XRCC1G28152A polymorphism was associated with a better survival in NSCLC patients following platinum-based chemotherapy [9, 10].
Thus, we hypothesised that a cancer patient’s inherited genotype for the GSTP1 and the XRCC1 gene may influence his or her survival, but evidence for a role of these genetic differences in lung cancer patients is limited. We investigated response rate and survival in relation to GSTP1 and XRCC1 genotypes in advanced NSCLC patients treated with platinum-based doublets as first-line chemotherapy.
Patients and methods
Patient selection
158 patients with histologically confirmed NSCLC were enrolled from October 2008 to February 2010 and were treated at Cancer Treatment Centre in the Affiliated Hospital of Qingdao University Medical College (China). In order to avoid the confounding effect of differences in outcome resulting from clinical stage, only NSCLC patients in stage IV were included in the analysis, and only patients with platinum-based doublets as first-line chemotherapy were recruited. There were 116 patients eligible for the study. The 116 patients, who were all Chinese Han people, also met the following prerequisites: (1) all patients had bi-dimensionally measurable disease with CT or MRI; (2) patients’ performance status was evaluated according to Eastern Cooperative Oncology Group (ECOG) criteria, and each patient’s ECOG status was not greater than 2; (3) each participant’s haemogram, hepatic function, renal function and electrocardiogram were normal before chemotherapy. All the 116 patients signed an informed consent form before entering the study.
Chemotherapy regimens
All patients had received platinum-based doublets as first-line chemotherapy as shown in table 1. Concrete dosage: DDP (cisplatin) 75 mg/m2on day 1; CBP (carboplatin) AUC = 4–5 g on day 1; DOC (docetaxel) 75 mg/m2 on day 1 (kept for 1h); GEM (gemcitabine) 1,250 mg/m2 on day 1 and day 8; NVB (vinorelbine) 25 mg/m2 on day 1 and day 8; PEM (pemetrexed) 500 mg/m2on day 1. All chemotherapeutic drugs were given intravenously, and the treatment cycles were repeated every 3–4 weeks.
Clinical evaluation
Patient response to treatment was determined after 2–3 cycles according to the Response Evaluation Criteria In Solid Tumours (RECIST) criteria [11]. In order to analyse the association between genotype and response to chemotherapy, patients with complete response (CR) and partial response (PR) were determined as “responders”. Patients with stable disease (SD) and progressive disease (PD) were referred to as “non-responders”. Time to progression (TTP) was calculated from the time of initial treatment to disease progression.
DNA isolation and genotyping
Two millilitres of peripheral whole blood samples were obtained from each patient before chemotherapy for DNA isolation and genotype determination. DNA was extracted from these samples using Whole Blood DNA Extraction Kit (Spin-column, Beijing BioTeKe Corporation). The genetic polymorphisms of GSTP1A313G and XRCC1G28152A were analysed by DNA sequencing methods. The primer sequences were as follows: (1)GSTP1gene,F:5’-GGAGTGGAGGAAACTGAGACC-3’,R:5’-GCAATAAGGGTGCAGGTTGT-3’;(2)XRCC1gene,F:5’-GCCCCTCAGATCACACCTAA-3’,R:5’-ACCGGGACTCACTTTGAATG-3’. Briefly, the 15 μl polymerase chain reaction (PCR) mixture contained 10×Buffer (Mg2+) 1.5 μl, PrimerF 0.3 μl, PrimerR 0.3 μl, dNTP 0.4 μl, Taq enzyme 0.4 μl and ddH2O 12.1 μl. Reactions were incubated at 94 °C for 2 min, then denatured at 94 °C for 30s, followed by 35 cycles of 58–60 °C for 45s and 72 °C for 50s, and extended at 72 °C for 7 min. The final products were purified using MultiScreen PCR 96-Well Plate (Millipore). Then ABI 3730XL sequencer (Applied Biosystems, USA) was used to determine the genotypes. Thirty randomly selected DNA samples were sequenced twice for confirmation and the results were 100% concordant.
Statistical analysis
Statistical analysis was performed using SPSS Software Package Version 13.0 (SPSS Inc., Chicago, IL, USA). Before assessing clinical associations, deviations from Hardy–Weinberg equilibrium for each SNP genotype were assessed using the Pearson χ2test. Demographic and clinical information was compared across genotype, also using Pearson χ2 tests. Contingency tables and Pearson χ2 tests were applied to test the significance of differences between genotypes and clinical response. We performed multivariate (logistic regression) analysis of the various contributing factors for response, such as gender, tumour histology, chemotherapy regimens, tumour differentiation and genetic polymorphisms. Odds ratios (OR) and their 95% confidence intervals (CI) were calculated using the binary logistic regression model. The genotype effect on patient’s survival was analysed using the Kaplan-Meier method. Comparison of survival curves was made using the Log-rank test. Hazard ratios (HRs) were determined using the Cox proportional hazard model. All p values reported were two-sided, and a probability level of less than 0.05 was considered statistically significant.
|
Table 1: Patient clinicopathological characteristics and chemotherapy regimens (n = 111). |
|
Characteristics
|
No. of Patients
|
%
|
| Age
≤60
>60 |
75
36 |
67.57
32.43 |
| Gender
Male
Female |
67
44 |
60.36
39.64 |
| Histology
Adenocarcinoma
Squamous cell carcinoma
Others |
53
47
11 |
47.75
42.34
9.91 |
| Differentiation
Well
Moderate
Poor |
17
54
40 |
15.31
48.65
36.04 |
| Performance status*
0
1
2 |
46
45
20 |
41.44
40.54
18.02 |
| Chemotherapy regimens
DDP/CBP+DOC
DDP/CBP+GEM
DDP/CBP+NVB
DDP/CBP+PEM |
44
37
21
9 |
39.64
33.33
18.92
8.11 |
| * Performance status was classified according to Eastern Cooperative Oncology Group (ECOG) criteria. |
Results
Patient characteristics
Finally, 111 of 116 patients enrolled in the study were eligible for data analysis. Three participants were withdrawn from the study because of failure of DNA extraction or genotyping, and two patients refused further chemotherapy due to unacceptable toxicity after one cycle of chemotherapy. The main characteristics of the 111 patients are shown in table 1. The median age was 57 years old (range 42–71 years).
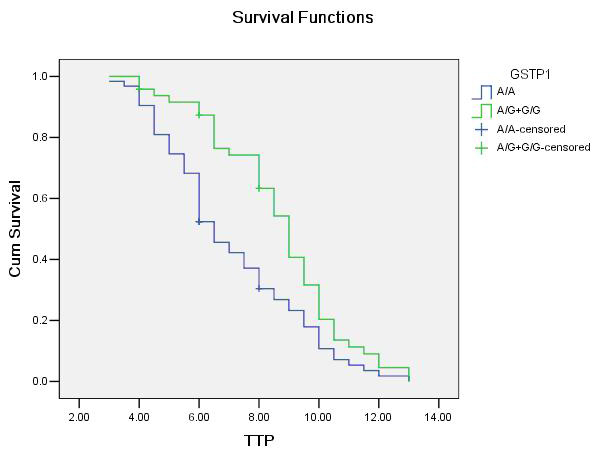
Figure 1
Association between GSTP1A313G genotypes (A/G and G/G as a combined group) and time to progression (TTP) in patients with advanced NSCLC receiving platinum-based chemotherapy. The vertical hash marks denote the time of last follow up for those patients lost to follow …up.
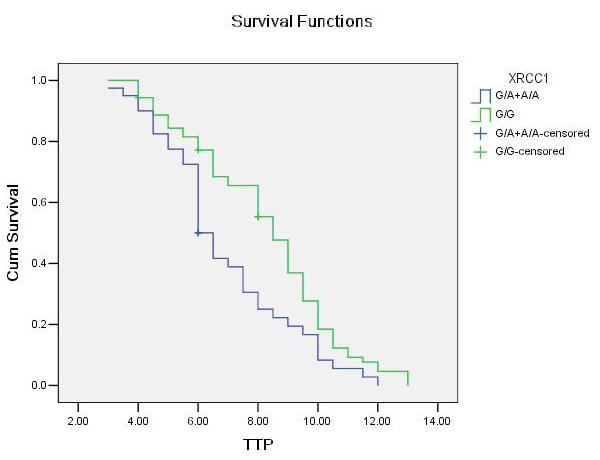
Figure 2
Association between XRCC1G28152A genotypes (G/A and A/A as a combined group) and time to progression (TTP) in patients with advanced NSCLC receiving platinum- based chemotherapy. The vertical hash marks denote the time of last follow up for those patients lost to follow up.
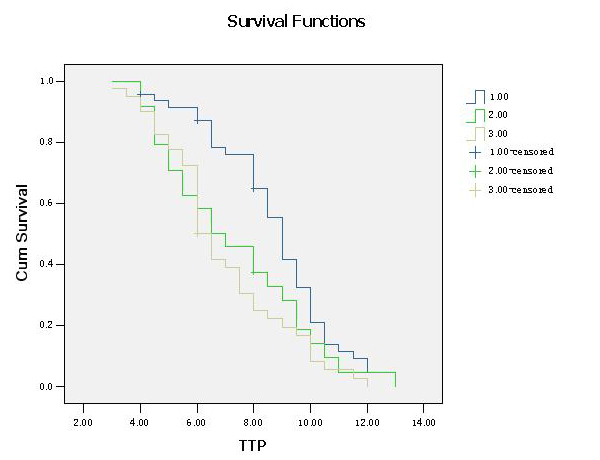
Figure 3
Association between the combination of GSTP1A313G and XRCC1G28152A polymorphisms (group 1: GSTP1 A/G+G/G and XRCC1 G/G genotype, group 2: GSTP1 A/G+G/G and XRCC1 G/A+A/A genotype, group 3: GSTP1 A/A and XRCC1 G/A+A/A genotype) and time to progression (TTP) in patients with advanced NSCLC receiving platinum- based chemotherapy. The vertical hash marks denote the time of last follow up for those patients lost to follow up.
Allele frequencies
The DNA sequencing analysis of the polymorphism located at GSTP1A313G showed that 63 (56.75%) were homozygous for A/A genotype, 40 (36.04%) were heterozygous for A/G genotype and 8 (7.21%) were homozygous for G/G genotype. The frequencies for A and G alleles were 74.77% and 25.23%, respectively. Owing to small sample size of G/G genotype, we integrated them with A/G genotype. For XRCC1G28152A, 71 (63.96%) were homozygous for G/G genotype, 34 (30.63%) were heterozygous for G/A genotype and 6 (5.41%) were homozygous for A/A genotype. The frequencies for G and A alleles were 79.28% and 20.72%, respectively. Also, we integrated the A/A genotype with G/A genotype. Genotype frequencies for GSTP1 and XRCC1 polymorphisms were both found to be in Hardy-Weinberg equilibrium (GSTP1: χ2 = 0.082, P = 0.960; XRCC1: χ2 = 0.155, P = 0.925). No associations were detected between genotype and age, gender, histological type or tumour differentiation (P >0.05).
Treatment response and genotype
Of the 111 patients, 35 patients experienced partial response (PR) whereas 28 showed stable disease (SD) and 48 experienced progressive disease (PD). The total chemotherapy efficiency was 31.53% (35/111).
GSTP1 genotype and treatment response
As shown in table 2, the GSTP1A313G polymorphism was significantly associated with response to platinum-based chemotherapy (χ2 = 8.013, P = 0.005). Patients with at least one variant allele (A/G+G/G) were more likely to respond to platinum-based chemotherapy compared with those with the homozygous wild genotype (A/A). Multivariate analysis adjusted for age, gender, tumour histology, tumour differentiation and chemotherapy regimens revealed that the GSTP1A313G polymorphism was independently associated with an objective response to platinum-based chemotherapy (OR, 3.961; 95%CI, 1.531–10.245; P = 0.005).
XRCC1 genotype and treatment response
For XRCC1G28152A genotype, patients with the homozygous wild genotype (G/G) had a higher response rate compared with those with at least one variant allele genotypes (G/A+A/A), and the difference was significant (χ2 = 7.916, P = .005) as shown in table 2. Multivariate analysis adjusted for age, gender, tumour histology, tumour differentiation and chemotherapy regimens showed that the XRCC1G28152A polymorphism was independently associated with response to platinum-based chemotherapy (OR, 3.700; 95%CI, 1.307–10.473; P = 0.014).
The combination of GSTP1A313G and XRCC1G28152A polymorphisms and treatment response
Based on the analysis results above, we defined GSTP1 A/G+G/G genotypes and XRCC1 G/G genotype as favourable genotypes and other genotypes as risk genotypes. It was notable that there were no individuals with four variant alleles, that is, no patients had GSTP1 G/G genotype and XRCC1 A/A genotype simultaneously. Chemotherapy efficiency of patients with GSTP1 A/G+G/G and XRCC1 G/G genotype (group 1), GSTP1 A/G+G/G and XRCC1 G/A+A/A genotype (group 2), GSTP1 A/A and XRCC1 G/A+A/A genotype (group 3) were 46.81% (22/47), 29.17% (7/24) and 15.00% (6/40) respectively. The difference was significant (χ2 = 10.207, P = 0.006). Multivariate analysis adjusted for age, gender, tumour histology, tumour differentiation and chemotherapy regimens revealed that the combination of GSTP1A313G and XRCC1G28152A polymorphisms was independently associated with response rate to platinum-based chemotherapy (P = 0.009), as shown in table 3.
Time to progression (TTP) and genotype
The median follow up time was 8.0 months (range 3.0 to 13.0 months). Overall, the median TTP was 8.0 (7.173 to 8.827) months for 105 patients. Until the end of follow up, all patients had disease progression and 6 patients who were lost to follow up were treated as censored data.
GSTP1 genotype and TTP
Patients with at least one variant genotype of the GSTP1 gene (A/G+G/G) had longer TTP in our study. Individuals with the homozygous wild genotype (A/A) had a median TTP of 6.5 (5.785 to 7.215) months, whereas those with at least one variant genotypes (A/G+G/G) had a median TTP of 9.0 (8.365 to 9.635) months (log-rank test, P = 0.004; table 4; fig. 1). In the Cox proportional hazards model, after adjusting for age, gender, tumour histology, tumour differentiation and chemotherapy regimens as an indicator variable, we found that the hazard ratio (HR) was higher for individuals with homozygous wild A/A genotypes (HR, 1.852; 95%CI, 1.185–2.893; P = 0.007) compared with A/G+G/G genotypes.
XRCC1 genotype and TTP
For XRCC1 genotype, individuals with the homozygous wild genotype (G/G) had a median TTP of 8.5 (7.857 to 9.143) months, whereas those with at least one variant genotype (G/A+A/A) had a median TTP of 6.0 (5.497 to 6.503) months (log-rank test, P = 0.007; table 4; fig. 2). In the Cox proportional hazards model, after adjusting for age, gender, tumour histology, tumour differentiation and chemotherapy regimens as an indicator variable, we found that the hazard ratio (HR) was higher for individuals with G/A+A/A genotypes (HR, 1.768; 95%CI, 1.143–2.734; P = 0.010) compared with wild genotypes (G/G).
The combination of GSTP1A313G and XRCC1G28152A polymorphisms and TTP
The median TTP of patients in group 1, group 2, group 3 were 9.000 (8.475 to 9.525) months, 6.500 (4.580 to 8.420) months and 6.000 (5.497 to 6.503) months respectively. The difference was significant (log-rank test, P = 0.007; table 5; fig. 3). In the Cox proportional hazards model, after adjusting for age, gender, tumour histology, tumour differentiation and chemotherapy regimens as an indicator variable, we found that the combination of GSTP1A313G and XRCC1G28152A polymorphisms was independently associated with TTP of the patients (P = 0.012), and the hazard ratio (HR) was significantly higher for individuals in group 3 (HR, 2.084; 95%CI, 1.282–3.390; P = 0.003) compared with patients in group 1.
|
Table 2: GSTP1 and XRCC1 polymorphism and chemotherapy response (n = 111). |
|
Genotype
|
Cases
|
Clinical response
|
Χ2-test
|
logistic regression
|
|
CR+PR
|
SD+PD
|
χ2
|
P
|
OR(95%CI)
|
P
|
| GSTP1
A/G+G/G
A/A |
48
63 |
22
13 |
26
50 |
8.013 |
0.005 |
1.000
3.961(1.531–10.245) |
0.005 |
| XRCC1
G/G
G/A+A/A |
71
40 |
29
6 |
42
34 |
7.916 |
0.005 |
1.000
3.700(1.307–10.473) |
0.014 |
|
Table 3: The combination of GSTP1 and XRCC1 polymorphisms and chemotherapy response (n = 111). |
|
Genotype
|
Cases
|
Clinical response
|
Χ2-test
|
logistic regression
|
|
CR+PR
|
SD+PD
|
χ2
|
P
|
OR(95%CI)
|
P
|
| Group 1
Group 2
Group 3 |
47
24
40 |
22
7
6 |
25
17
34 |
10.207
|
0.006 |
1.000
3.421(0.943–12.409)
5.516(1.775–17.140) |
0.061
0.003 |
|
Table 4: GSTP1 and XRCC1 polymorphism and TTP (n = 105). |
|
Genotype
|
Cases
|
Median TTP
(95%CI)
|
log-rank test
|
Cox regression analysis
|
|
χ2
|
P
|
OR(95%CI)
|
P
|
| GSTP1
A/G+G/G
A/A |
45
60 |
9.0(8.365–9.635)
6.5(5.785–7.215) |
8.233 |
0.004 |
1.000
1.852(1.185–2.893) |
0.007 |
| XRCC1
G/G
G/A+A/A |
67
38 |
8.5(7.857–9.143)
6.0(5.497–6.503) |
7.225 |
0.007 |
1.000
1.768(1.143–2.734) |
0.010 |
|
Table 5: The combination of GSTP1 and XRCC1 polymorphisms and TTP (n = 105). |
|
Genotype
|
Cases
|
Median TTP
(95%CI)
|
log-rank test
|
Cox regression analysis
|
|
χ2
|
P
|
OR(95%CI)
|
P
|
| Group 1
Group 2
Group 3 |
44
23
38 |
9.0(8.475–9.525)
6.5(4.580–8.420)
6.0(5.497–6.503) |
9.839 |
0.007 |
1.000
1.631(0.902–2.950)
2.084(1.282–3.390) |
0.106
0.003 |
Discussion
Nowadays, pharmacogenetics is becoming an increasingly intriguing field in the study of cancer chemotherapy. There is a growing body of evidence suggesting that genetic polymorphisms in genes involved in metabolism, signalling, DNA-repair and cellular response pathways may contribute to inter individual variability of drug response and toxicity [12, 13], but little has been successfully translated to the clinical setting. Given that the diagnosis of advanced cancers is mainly based on a small needle biopsy sample, using tumour tissue to acquire further predictive or prognostic information may be difficult. Thus, assessing germline genetic polymorphisms from peripheral venous blood either as prognostic or predictive markers becomes much more appealing, especially in the advanced cancer setting.
In our study, we attempted to examine whether polymorphisms of GSTP1 and XRCC1 genes related to the metabolism of cisplatin and DNA repair could be used as predictors of the clinical outcome of patients with advanced NSCLC treated with platinum-based chemotherapy. We found that the GSTP1 and XRCC1 genetic polymorphisms, both alone and in combination, are associated with chemosensitivity and clinical outcome in this patient population.
Glutathione S-transferases are crucial to the cell defence system, and these phase II detoxification enzymes are involved in the detoxification of a variety of chemotherapeutics including platinum. Glutathione S-transferases P1 (GSTP1), located on chromosome 11q13 in humans, is expressed in many human epithelial tissues and is the most abundant GST isoform in the lung. A313G in exon 5 is one of the common single nucleotide polymorphisms in GSTP1 that lead to amino acid substitutions. In addition, studies suggest that the variant genotype results in diminished enzymatic activity. Thus, individuals with variant GSTP1 genotypes may possess increased susceptibility to cancer but good responses to chemotherapy due to decreased detoxification of carcinogens and chemotherapeutic agents. Numerous epidemiologic studies have studied possible associations between GSTP1 polymorphisms and the risk of lung cancer, often with controversial results [14–17]. Clinical studies have also implicated GSTP1 as a predictive marker for clinical outcome in patients with cancer treated with platinum-based chemotherapy [18–20]. Sun et al. [21] found that the A→G change of GSTP1A313G polymorphism significantly increased platinum-based chemotherapy response in advanced NSCLC patients. Our results indicate that A/G and G/G genotypes of codon 105 polymorphism of GSTP1 gene are favourable genotypes for chemotherapy response and survival of NSCLC patients receiving platinum-based chemotherapy and are consistent with previous study reports [18, 20, 21].
Growing evidence has suggested that single nucleotide polymorphisms (SNPs) in DNA repair genes can affect repair efficiency [22, 23]. Suboptimal DNA repair may lead to the decreased removal of platinum-DNA adducts and may therefore increase cancer risk [24, 25] and clinical response to platinum therapy [26]. The X-ray repair cross complementing group 1 (XRCC1) gene is a member of the base excision repair (BER) pathway. The XRCC1 gene product plays an important role in the BER pathway by acting as a scaffold for other DNA repair proteins, such as DNA polymerase β and DNA ligase III [27]. Genetic polymorphisms of the XRCC1 gene have been identified at C26304T (codon 194,exon 6) and G28152A (codon 399,exon 10), and these polymorphisms may confer different activity. Carriers of the XRCC1-399 A/A variant allele have been shown to have higher levels of DNA adducts [28] and tobacco-related DNA damage [29, 30] and an increased risk of grade 3 or 4 gastrointestinal toxicity when treated with first line cisplatin-based chemotherapy [31]. Sarada Gurubhagavatula et al. [32] reported that advanced NSCLC patients with XRCC1-399 variant genotype (A/A) were associated with shorter overall survival. Our results found that G/G genotype of codon 399 are favourable genotypes for chemotherapy response and survival of NSCLC patients receiving platinum-based chemotherapy.
Furthermore, it highlights the fact that combined GSTP1 and XRCC1 gene polymorphisms are involved in glutathione metabolic and DNA repair pathways and we found that these two favourable genotypes were significantly associated with a good treatment outcome. This causes us to believe that in future studies we should focus on different mechanisms related to platinum agent resistance simultaneously. It is also interesting that the median TTP of patients in group 2 (GSTP1 A/G+G/G and XRCC1 G/A+A/A genotype) was shorter than patients with GSTP1 A/G+G/G genotype alone (9.0 months vs 6.5 months). We may infer that the more favourable response of GSTP1 A/G+G/G carrier is partially neutralised by the presence of XRCC1 G/G+A/A, the unfavourable genotype. This phenomenon must be confirmed by subsequent research.
However, some conflicting reports exist in the literature [33–36]. Due to several different aspects of study design, including size of the study populations, observation of NSCLC patients in particular stages or of all lung cancers, length of follow up and choice of different end points, discordant results may appear. Other possible explanations may be attributed to some other potential factors that might affect the clinical outcome to chemotherapy, such as ethnic background, geographic pattern or other potential genetic factors. In addition, the limitations of our study must be acknowledged. Due to the relatively small sample size, we intend to expand our sample size in subsequent studies to acquire further validation and to concentrate on overall survival, the most objective outcome in our study population.
Based on early research and our study, we speculated that the GSTP1A313G variant genotype (A/G+G/G) and XRCC1G28152A wild genotype (G/G) are associated with improved clinical outcome among patients with advanced NSCLC treated with platinum-based chemotherapy. Further studies evaluating platinum-based chemotherapy and overall survival data are needed to validate these findings and to determine the prognostic significance of GSTP1 and XRCC1 genotype in patients with advanced NSCLC. Despite the limitations mentioned above, the data from our study contribute significant information on the prognostic and predictive value of these polymorphisms and may prove to be useful tools whilst working towards individualising NSCLC treatment strategies.
References
1 Yang L, Parkin DM, Ferlay J, Li L, Chen Y. Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer Epidemiol Biomarkers Prev. 2005;14(1):243–50.
2 Spiro SG, Silvestri GA. The treatment of advanced non-small cell lung cancer. Curr Opin Pulm Med. 2005;11(4):287–91.
3 Kelly K, Crowley J, Bunn PA Jr, Presant CA, Grevstad PK, Moinpour CM, et al. Randomized phase III trial of paclitaxel plus carboplatin versus vinorelbine plus cisplatin in the treatment of patients with advanced non – small-cell lung cancer: a Southwest Oncology Group trial. J Clin Oncol. 2001;19(13):3210–8.
4 Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–8.
5 Fuertes MA, Castilla J, Alonso C, Perez JM. Cisplatin biochemical mechanism of action: from cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr Med Chem. 2003;10(3):257–66.
6 Peklak-Scott C, Smitherman PK, Townsend AJ, Morrow CS. Role of glutathione S-transferase P1-1 in the cellular detoxification of cisplatin. Mol Cancer Ther. 2008;7(10):3247–55.
7 Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19(2):275–80.
8 Booton R, Ward T, Heighway J, Ashcroft L, Morris J, Thatcher N. Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol. 2006;1(7):679–83.
9 Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, et al. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22(13):2594–601.
10 Giachino DF, Ghio P, Regazzoni S, Mandrile G, Novello S, Selvaggi G, et al. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin Cancer Res. 2007;13(10):2876–81.
11 Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–16.
12 Allen WL, Johnston PG. Role of genomic markers in colorectal cancer treatment. J Clin Oncol. 2005;23(20):4545–52.
13 Yamayoshi Y, Iida E, Tanigawara Y. Cancer pharmacogenomics: international trends. Int J Clin Oncol. 2005;10(1):5–13.
14 Jourenkova-Mironova N, Wikman H, Bouchardy C, Voho A, Dayer P, Benhamou S, et al. Role of glutathione S-transferase GSTM1, GSTM3, GSTP1 and GSTT1 genotypes in modulating susceptibility to smoking-related lung cancer. Pharmacogenetics. 1998;8(6):495–502.
15 To-Figueras J, Gene M, Gomez-Catalan J, Pique E, Borrego N, Carrasco JL, et al. Genetic polymorphism of glutathione S-transferase P1 gene and lung cancer risk. Cancer Causes Control. 1999;10(1):65–70.
16 Harris MJ, Coggan M, Langton L, Wilson SR, Board PG. Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients. Pharmacogenetics. 1998;8(1):27–31.
17 Wang Y, Spitz MR, Schabath MB, Ali-Osman F, Mata H, Wu X. Association between glutathione S-transferase p1 polymorphisms and lung cancer risk in Caucasians: a case-control study. Lung Cancer. 2003;40(1):25–32.
18 Stoehlmacher J, Park DJ, Zhang W, Groshen S, Tsao-Wei DD, Yu MC, et al. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2002;94(12):936–42.
19 Ott K, Lordick F, Becker K, Ulm K, Siewert J, Hofler H, et al. Glutathione-S-transferase P1, T1 and M1 genetic polymorphisms in neoadjuvant-treated locally advanced gastric cancer: GSTM1-present genotype is associated with better prognosis in completely resected patients. Int J Colorectal Dis. 2008;23(8):773–82.
20 Stoehlmacher J, Park DJ, Zhang W, Yang D, Groshen S, Zahedy S, et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91(2):344–54.
21 Sun N, Sun X, Chen B, Cheng H, Feng J, Cheng L, et al. MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2010;65(3):437–46.
22 Abdel-Rahman SZ, El-Zein RA. The 399Gln polymorphism in the DNA repair gene XRCC1 modulates the genotoxic response induced in human lymphocytes by the tobacco-specific nitrosamine NNK. Cancer Lett. 2000;159(1):63–71.
23 Au WW, Navasumrit P, Ruchirawat M. Use of biomarkers to characterize functions of polymorphic DNA repair genotypes. Int J Hyg Environ Health. 2004;207(4):301–13.
24 Auranen A, Song H, Waterfall C, Dicioccio RA, Kuschel B, Kjaer SK, et al. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117(4):611–8.
25 Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(12):1513–30.
26 Bosken CH, Wei Q, Amos CI, Spitz MR. An analysis of DNA repair as a determinant of survival in patients with non-small-cell lung cancer. J Natl Cancer Inst. 2002;94(14):1091–9.
27 Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst). 2003;2(9):955–69.
28 Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res. 1999;59(11):2557–61.
29 Duell EJ, Wiencke JK, Cheng TJ, Varkonyi A, Zuo ZF, Ashok TD, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis. 2000;21(5):965–71.
30 Lei YC, Hwang SJ, Chang CC, Kuo HW, Luo JC, Chang MJ, et al. Effects on sister chromatid exchange frequency of polymorphisms in DNA repair gene XRCC1 in smokers. Mutat Res. 2002;519(1-2):93–101.
31 Wang Z, Xu B, Lin D, Tan W, Leaw S, Hong X, et al. XRCC1 polymorphisms and severe toxicity in lung cancer patients treated with cisplatin-based chemotherapy in Chinese population. Lung Cancer. 2008;62(1):99–104.
32 Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, et al. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22(13):2594–601.
33 Sweeney C, Nazar-Stewart V, Stapleton PL, Eaton DL, Vaughan TL. Glutathione S-transferase M1, T1, and P1 polymorphisms and survival among lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2003;12(6):527–33.
34 Lu C, Spitz MR, Zhao H, Dong Q, Truong M, Chang JY, et al. Association between glutathione S-transferase pi polymorphisms and survival in patients with advanced nonsmall cell lung carcinoma. Cancer. 2006;106(2):441–7.
35 Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, et al. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10(2):118–23.
36 Ada AO, S CK, Hancer F, Bilgen S, Suzen SH, Alpar S, et al. CYP and GST polymorphisms and survival in advanced non-small cell lung cancer patients. Neoplasma. 2010;57(6):512–21.


