
Figure 1
Algorithm to decide whether to screen/isolate for MRSA at admission.
DOI: https://doi.org/10.4414/smw.2011.13217
MRSA (Methicillin-resistant Staphylococcus aureus) is one of the most important nosocomial pathogens world-wide causing significant morbidity (blood stream infection, skin and soft-tissue infection, pneumonia) and mortality (twice as high as with Methicillin-susceptible Staphylococcus aureus (MSSA), at least in blood stream infection) [1–3]. Once colonised with MRSA, 11–25% of patients in acute-care facilities and 3–15% in chronic-care facilities will subsequently develop infections [4–7]. MRSA-colonised patients are much more likely to develop infection during a one-year follow-up than MSSA- or non colonised patients (19–25% versus 1.5–2.0%) [8].

Figure 1
Algorithm to decide whether to screen/isolate for MRSA at admission.
MRSA prevalence varies considerably among European countries ranging from <1% in Sweden, Norway and the Netherlands to >50% in Portugal [9]. With a level of 11.5% in 2008, the MRSA prevalence in Switzerland was lower than in most of its neighbouring countries (Austria: 8.2%, Germany: 19.5%, France: 24.5%, Italy: 33.5%). Within Switzerland, it was as high as 17.3% in Western Switzerland and as low as 2.5% in the area around St. Gallen (MRSA rate in clinical isolates) [10].
In 2007, a doubling of the number of newly diagnosed MRSA patients was observed at the Cantonal Hospital in St. Gallen, which was mainly attributable to a rise of one MRSA-genotype known to be prevalent in local chronic care facilities. Guidelines for MRSA admission screening/pre-emptive isolation, previously mainly focussed on admissions from foreign hospitals and MRSA high prevalence regions of Switzerland (Geneva, Vaud, Ticino), were adapted accordingly. Following a new algorithm (fig. 1), patients were risk stratified into screening and isolation, only screening or no screening at all – depending on epidemiological factors (stay in a foreign hospital, a Swiss hospital with known high MRSA prevalence, or a chronic care facility) and personal risk factors (wound, tracheostoma, urinary catheter, intravenous drug use (IVDU)). MRSA admission screening included nasal, throat and axillary/inguinal swabs, supplemented by wound swabs, urine and respiratory secretion whenever appropriate (conventional culture).
Many hospitals practice some sort of MRSA admission screening. It is generally accepted as one of a package of measures in MRSA prevention [11], because early diagnosis of MRSA can prevent its spreading within a hospital [5, 12, 13]. However, the specific value has been discussed controversially [14, 15], particularly in low-prevalence regions, where the yield may be extremely low [16]. Some countries, such as the United Kingdom, have introduced a mandatory MRSA admission screening [17], while others restrict screening to special wards (e.g. intensive care unit) [18] or high-risk patients [13]. Occasionally, even weekly follow-up screening during hospitalisation is propagated [18, 11]. A large Swiss study failed to demonstrate an effect of universal admission screening on nosocomial MRSA-infection rates [19], and pre-emptive isolation is not without side effects, particularly with regard to patient safety [14, 20, 21]. Frequently, the number of screening sites and the method of MRSA-detection (conventional culture, chromogenic agar, PCR) is also a point of discussion [11].
Generalisability of study results is often limited due to the diversity of settings and mostly multiple interventions at the same time [14, 22]. Thus, we considered it necessary to prospectively evaluate our new MRSA admission screening/pre-emptive isolation strategy during a one-year surveillance. The following issues were addressed:
– total number of patients screened and pre-emptively isolated following the new algorithm,
– proportion of screened/isolated patients actually MRSA-positive,
– number needed to screen and number needed to isolate,
– proportion of newly diagnosed MRSA patients detected by admission screening and clinical sample during hospitalisation, respectively, as well as
– the value of different screening sites.
Furthermore, prevalence of known risk factors for MRSA colonisation, MRSA-genotypes and delay of MRSA diagnosis were compared between patients newly MRSA-diagnosed by admission screening and clinical sample during hospitalisation, respectively.
The surveillance was performed from 04/2008 to 03/2009 in patients admitted to the tertiary-care hospital in St. Gallen (700 beds) and 10 affiliated public hospitals (890 beds) in Eastern Switzerland. Except for known MRSA carriers, all patients who underwent admission screening in accordance with the guidelines (fig. 1) and all MRSA patients newly detected by clinical sample were included.

Figure 2
Performance of the new guidelines for MRSA admission screening and pre-emptive isolation during the one-year surveillance 04/2008–03/2009.
A+B) MRSA rate among patients screened (A)/pre-emptively isolated (B) at admission and number needed to screen (A)/isolate (B)
C+D) Proportion of newly diagnosed MRSA patients detected by screening (C)/ pre-emptively isolated (D)
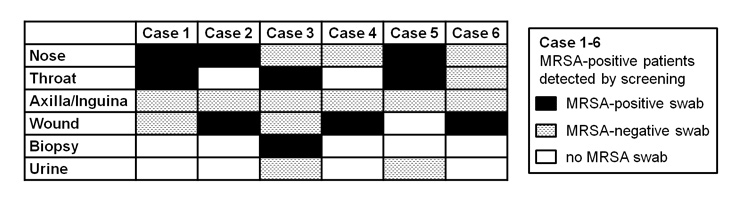
Figure 3
Positive screening sites in case of positive MRSA admission screening.
Swabs of axilla/inguina did not increase the sensitivity of the admission screening.
There was no universal MRSA admission screening by swabs, but the decision of whether to screen/pre-emptively isolate a patient was based on a risk stratification: high risk for MRSA carriage – screening and pre-emptive isolation, moderate risk – only screening, no isolation and low risk – neither screening nor isolation. As depicted in figure 1, the screening guidelines considered epidemiological factors (stay in a foreign hospital, a Swiss hospital with a high known MRSA prevalence, or a chronic care facility within the past six months) and personal risk factors (wound, tracheostoma, urinary catheter and IVDU (intravenous drug use)). Patients from chronic care facilities (old people’s or nursing home, rehabilitation clinic) and IVDUs were only screened if additional risk factors (suppurating wound, tracheostoma/intubation, urinary catheter) were present. In contrast, all patients hospitalised in a foreign hospital or a hospital in a MRSA high prevalence region of Switzerland (Geneva, Vaud, Ticino) for >24 h within the past six months were screened and those with additional risk factors were pre-emptively isolated awaiting screening results.
The MRSA admission screening consisted of nasal, throat and axillary/inguinal swabs as well as wound swabs, urine in the case of a urinary catheter and respiratory secretion provided the patient was intubated or a tracheostoma was present. The swabs (COPAN 108C) were moistened with sterile 0.9% NaCl-solution (10 ml, Braun) before use.
Pre-emptively isolated patients were placed in a single room. Health care workers wore gown and gloves during direct patient contact and surgical masks if exposure to respiratory secretion was to be expected (e.g., coughing patient). Pre-emptive isolation measures were abolished as soon as the admission screening turned out to be negative.
For MRSA detection, a conventional culture with cefoxitin diffusion disc test (according to Clinical and Laboratory Standards Institute (CLSI) guidelines) was used [23, 24]. The turn-around time was 3–4 days.
MRSA-isolates were epidemiologically genotyped by pulsed-field gel electrophoresis in the Laboratory of Hospital Epidemiology, University Hospital of Zurich, as published in Fleisch et al. [25].
The Institute of Clinical Microbiology routinely informs the Department of Infectious Diseases about every positive MRSA result by telephone call or mail. In addition, during the one-year surveillance period, it provided a copy of all MRSA screenings irrespective of the result.
Data on risk factors were prospectively collected by reviewing electronic medical charts. In the case of MRSA positive patients, additional information was available, because the infection control team contacted the patient directly on the ward within the scope of its duties.
The following issues were addressed in the one-year prospective evaluation of the new MRSA admission screening/pre-emptive isolation strategy:
– total number of patients screened and pre-emptively isolated following the new algorithm,
– proportion of screened/isolated patients actually MRSA-positive,
– number needed to screen and number needed to isolate,
– proportion of newly diagnosed MRSA patients detected by admission screening and clinical sample during hospitalisation, respectively, as well as
– the value of different screening sites.
Furthermore, prevalence of known risk factors for MRSA colonisation, proportion with the locally dominant MRSA-genotype 21 and delay of MRSA diagnosis (time from admission to MRSA swab as well as time from admission to MRSA diagnosis) were compared between patients newly MRSA-diagnosed by admission screening and clinical sample during hospitalisation, respectively.
The number needed to screen/isolate was calculated as the reciprocal of the proportion of screened/isolated patients actually MRSA-positive. 95%-confidence intervals were computed with Open Source Epidemiologic Statistics for Public Health, Version 2.3 (www.OpenEpi.com http://www.openepi.com/ ).
The prevalence of known risk factors for MRSA colonisation was compared between negative and positive screened patients and patients newly MRSA-diagnosed by clinical sample during hospitalisation, respectively. Statistical analyses were performed using StatView (version 5.0; SAS Institute Inc., Cary, NC). All reported p values are two-sided, and p values <0.05 were considered significant. Chi Square and Fisher’s Exact Test were used as appropriate.
No formal informed consent was obtained because the study was strictly observational and part of the institutional quality control activities.
Number of patients screened/pre-emptively isolated at admission and total number of newly diagnosed MRSA patients during the one-year surveillance
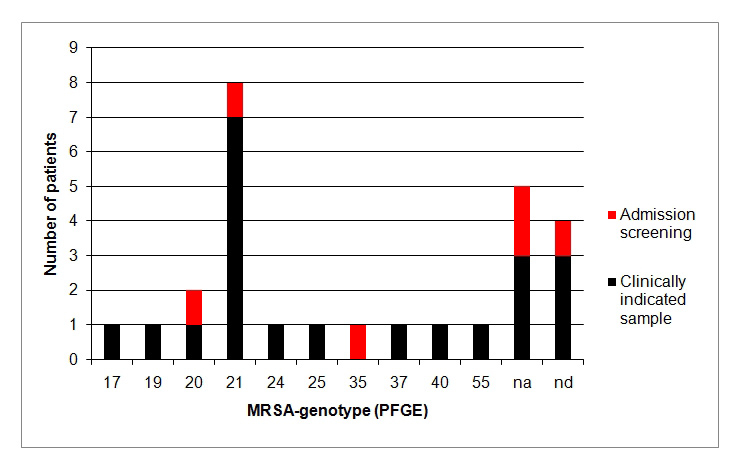
Figure 4
MRSA-genotyping by PFGE.
In four patients, MRSA-genotyping by PFGE (Pulsed-field gel electrophoresis) was not performed (nd = not determined). MRSA-genotypes of five patients were not attributable (na = not attributable), i.e. locally not known. MRSA-genotype 21 with known prevalence in local chronic care facilities predominated.

Figure 5
Time from admission to MRSA swab and MRSA diagnosis.
Following the algorithm depicted in figure 1, during one year, 161 patients were screened for MRSA at admission (111 in St. Gallen (0.4% of ~28,000 admissions/year in 2008) and 50 in the affiliated hospitals (0.125% of ~40,000 admissions/year in 2008)). About 20% of them (32/161) were pre-emptively isolated while awaiting screening results. During the same time period (04/2008–03/2009), MRSA was newly diagnosed in a total of 27 patients.
Patients hospitalised in the same room as newly diagnosed, and until then, not isolated MRSA-patients, were screened by swabs at day one and five after last contact. This routinely performed contact screening failed to identify further MRSA-positive patients as index or secondary cases (i.e. no nosocomial transmission observed).
MRSA rate among patients screened/pre-emptively isolated at admission and number needed to screen/isolate
A total of 6/161 (3.7%; 95% CI: 2–8%) MRSA admission screenings yielded positive results. Accordingly, the number needed to screen was 27 (95% CI: 13–67) (fig. 2A). Of the 32 pre-emptively isolated patients, two (6.3%; 95% CI: 1–19%) were actually MRSA-positive, resulting in a number needed to isolate of 16 (95% CI: 5–91) (fig. 2B).
Proportion of newly diagnosed MRSA patients detected by screening/pre-emptively isolated
Of the 27 newly diagnosed MRSA patients, six (22.2%; 95% CI: 10–41%) were detected by admission screening and 21 (77.8%; 95% CI: 59–90%) by clinically indicated samples (figure 2C). A total of 7.4% (95% CI: 1–22%) of them (2/27) had been pre-emptively isolated, which was two of the six MRSA patients detected by admission screening (fig. 2D).
All six patients diagnosed by MRSA admission screening were detected either by nasal, throat or wound swabs. Samples from axilla/inguina did not increase the sensitivity of the admission screening (no positive results) (fig. 3).
Among the 21 patients diagnosed by a clinically indicated sample, all – except for one patient with only positive urine – would have been detected by a screening consisting of nasal, throat and wound swabs. However, in this population, supplementary screening of axilla/inguina after MRSA diagnosis yielded positive results in 50% (data not shown).
Classical risk factors for MRSA like tracheostoma, intravenous drug use and dialysis were more frequent in negative versus positive screened patients. In contrast, the prevalence of wounds (skin lesions) and immunosuppression tended to be higher in positive versus negative screened patients. A wound, as a risk factor, was present in 80% of MRSA positive patients, no matter whether they were diagnosed by screening or a clinically indicated sample (table 1).
None of the MRSA patients diagnosed by clinical sample had been hospitalised in a foreign country within the last 6 months compared to 83.3% (5/6) of the positive screened patients, indicating good adherence to the guidelines to screen all patients who had stayed in a foreign hospital (table 1, fig. 1).
For 23/27 (85%) newly diagnosed MRSA patients, PFGE-genotype was determined (fig. 4). About one-third (8/23) of these patients had MRSA-genotype 21 (1/5 (20%) of positive screened and 7/18 (39%) of clinical diagnosed patients; no significant difference (p = 0.5)). In 22% (5/23) genotype was not attributable (i.e. locally unknown).
None of the eight MRSA-genotype 21-patients had been hospitalised in a foreign country, whereas three of them had been transferred from the University Hospital Zurich, a nursing home and a rehabilitation clinic, respectively.
A total of 98% (158/161) of all MRSA admission screenings, but only 57% (12/21) of swabs in clinically diagnosed MRSA patients were performed within the first 24 hours after admission. With a turn-around time of 3–4 days for conventional culture, this resulted in a median time from admission to MRSA diagnosis of 3 days (range 3–4 days) for admission screening and 5 days (range 2–21 days) for clinically indicated samples (fig. 5).
| Table 1:Risk factors in screened and clinically diagnosed patients. | |||
| MRSA screening | MRSA detection by clinical sample (n = 21) | ||
| Negative (n = 155) | Positive (n = 6) | ||
| Age (mean, 95%-CI) | 61.3 ± 2.9 | 60.8 ± 13.3 | 56.1 ± 8.6 |
| Gender, % female | 43.2 (67/155) | 33.3 (2/6) | 57.1 (12/21) |
| Personal RF % | |||
| Skin leason | 39.2 (47/120) | 83.3 (5/6)$ | 80.9 (17/21) |
| Tracheostoma | 10.5 (13/124) | 0 (0/6) | 5.3 (1/19) |
| Urine catheter | 32.2 (38/118) | 33.3 (2/6) | 21.1 (4/19) |
| PEG | 4.8 (6/124) | 0 (0/6) | 0 (0/19) |
| IVDU | 3.9 (5/127) | 0 (0/6) | 0 (0/18) |
| Immunosuppression | 1.7 (2/118) | 16.7 (1/6)§ | 11.1 (2/18) |
| Dialysis | 9.5 (12/126) | 0 (0/6) | 5.0 (1/20) |
| Diabetes | 13.6 (16/118) | 0 (0/6) | 26.3 (5/19) |
| Epidemiological RF % | |||
| Foreign hospital | 65.8 (75/114) | 83.3 (5/6) | 0 (0/18)* |
| CH high prevalence hospital | 9.5 (11/116) | 0 (0/6) | 0 (0/18) |
| Chronic care facility | 20.7 (25/121) | 16.7 (1/6) | 9.5 (2/21) |
| CI: Confidence Interval; RF: Risk factor; PEG: Percutaneous endoscopic gastrostomy; IVDU: Intravenous drug use, CH: Switzerland $ p = 0.08, § p = 0.13, Screening neg vs pos; *p <0.001, Screening vs clinical pos | |||
Results of the current one-year surveillance evaluating newly implemented guidelines for MRSA-admission screening and pre-emptive isolation raised several questions, which will be consecutively discussed in light of the available literature.
In the UK (MRSA rate 30% in 2008 [9]), MRSA screening has been mandatory for all elective and day case admissions since April 2009 and for all emergency admissions since April 2010. In the setting of universal admission screening in a 1095 bed hospital in Southampton (nose and groin in all, wounds and catheters when present; during one year >160,000 MRSA swabs on 51,855 individuals costing 600,000 Euros (chromogenic agar)), the total MRSA rate was 1.05% (0.5% for elective and 1.5% for emergency admissions), resulting in a number needed to screen of 95 (range 24 to >1006 depending on clinical speciality and admission type) [17].
In the Netherlands (1% MRSA rate), universal nasal MRSA admission screening in patients without risk factors yielded only 3 MRSA carriers in almost 10,000 patients (0.03%) [16], resulting in a number needed to screen of 3333 (95% CI: 1227–12,549). In our setting with a comparable low MRSA rate in isolates (2.5%), restricting microbiological MRSA screening and pre-emptive isolation to patients with a high risk of MRSA carriage (cf. algorithm fig. 1) resulted in significantly lower numbers needed to screen and to isolate (27 (95% CI: 13–67) and 16 (95%: 5-91), respectively). If MRSA admission screening by swabs had been applied to all 68,000 admissions irrespective of their risk factors and all 27 new MRSA cases had been detected that way, the number needed to screen would have been 2519 (95% CI: 1758–3705). Of course, it cannot be excluded that there were MRSA positive patients undetected throughout their hospital stay, who might have been diagnosed by a universal microbiological MRSA admission screening.
Thus, for the sake of cost effectiveness, individualised screening strategies seem to be more favourable in a low prevalence setting. However, the relatively high proportion of positive MRSA admission screenings (3.7% [6/161]) must be weighed against the fact that only 22% (6/27) of newly detected MRSA patients were diagnosed by admission screening. Data from the year after the study (04/2009–03/2010) confirmed that our screening strategy detected only one out of five newly diagnosed MRSA patients (35 new MRSA patients: 7/35 (20%, 95% CI: 9–36%) detected by admission screening and 28/35 (80%, 95% CI: 64–91%) by clinical sample).
Contact isolation (single room, patient contact with gown and gloves) is not without side effects (fewer health care worker visits, more non-infectious adverse events, more depression and anxiety, lower satisfaction with hospital care) [14, 20, 21]. Taking into account the huge number of unnecessary isolation days resulting from the low MRSA prevalence and the long turn-around time for screening tests (3–4d) in our setting, it did not seem to be ethical[26] and cost-effective to pre-emptively isolate all patients awaiting screening results.
According to the algorithm (fig. 1), only 19.9% (32/161) of screened patients were isolated, with 2 of them (6.3%) being MRSA positive (Number needed to isolate: 16). However, thus only 7.4% (2/27) of all newly diagnosed MRSA patients were pre-emptively isolated, who were two of the six MRSA patients detected by admission screening (fig. 2).
Compliance with standard precautions, particularly hand hygiene, determines the transmission risk while undetected MRSA positive patients are not contact isolated. With 80%, hand hygiene compliance was rather high in our setting (measured in accordance with the 5 WHO recommended indications in an observation study during autumn 2009, as described in Sax et al. [27]). Accordingly, contact screening, which was routinely performed for all newly diagnosed MRSA-patients not isolated until then, failed to identify further MRSA-positive patients as index or secondary cases (i.e. no nosocomial transmission observed) (data not shown).
The use of alternative screening tests (e.g., chromogenic agar media, PCR-based testing) might reduce the turn-around time from 3–4 days for standard culture to 24h [11], but would also raise additional costs challenging cost-effectiveness (costs for microbiological MRSA diagnostic in the Cantonal Hospital St. Gallen: negative culture: 25 CHF, positive culture: 70 CHF, PCR: 180 CHF; 1 CHF corresponds to 0.87 USD (exchange rate 21th May 2010)).
Cost effectiveness of PCR-based MRSA screening depends on the number of isolation days that can be saved [28]. In our setting, with only a small proportion of admitted patients screened and even a smaller proportion pre-emptively isolated (32 per year), the potential of saving money with a faster, but more expensive method is rather small. 60 isolation days (30x 2 days at 300 CHF for single room = 18,000 CHF) could have been saved, if MRSA screening results had been available two days earlier in the 30 pre-emptively isolated but MRSA negative screened patients. However, screening all 161 patients or only the 32 pre-emptively isolated patients with PCR, would have resulted in additional costs of about 25,000 and 5000 CHF per screening site, respectively.
With the current method (standard culture), the median time from admission to MRSA diagnosis was 3 days for patients diagnosed by admission screening and 5 days for those diagnosed by clinically indicated samples.
The sensitivity of nasal MRSA screening alone is 79% (NPV 95%), which can be increased to 98% by additional screening sites (throat, groin) (NPV 99%) [29]. Exclusive throat carriers would be missed by nasal screening only [30].
In our setting, all six patients diagnosed by MRSA admission screening would have been detected with a combination of nasal, throat and wound swabs, whereas none of the samples from axilla/inguina delivered positive results (fig. 3). Also, among the 21 patients diagnosed by a clinically indicated sample, all – except for one patient with only positive urine – would have been detected by screening the nose, throat and wound only, though supplementary screening of axilla/inguina after MRSA diagnosis yielded positive results in 50%. The small number of positive admission screenings prevented us from immediately removing axilla/inguina from the admission screening. In the following year (04/2009–03/2010), 5/7 patients with positive admission screening would have been detected by screening the nose, throat and wound only. However, 2/7 would have been missed being positive only in the axilla/inguina swab. Accordingly, up to now, we have not reduced the number of screening sites.
Prior guidelines for MRSA admission screening at the Cantonal Hospital, St. Gallen, only focussed on admissions from foreign hospitals and MRSA high prevalence regions of Switzerland (Geneva, Vaud, Ticino). However, the doubling of the number of newly diagnosed MRSA patients in 2007 was primarily attributable to a rise of the MRSA genotype 21 known to be prevalent in local chronic care facilities.
Extending MRSA-admission screening to IVDUs and chronic care facility residents with additional risk factors resulted in about 30% more screenings and 20% more positive screening results (i.e. six instead of five) (table 1). A total of 8/23 newly diagnosed MRSA patients with available genotype were MRSA genotype 21 (none of them hospitalised in a foreign country), but only one (transferred from a rehabilitation clinic) was detected by the adapted admission screening. Adherence with the new screening guidelines might have been a limiting factor, because at least one more MRSA genotype 21 patient, later diagnosed by a clinical sample, would have qualified for admission screening. In contrast, none of the MRSA patients diagnosed by clinical sample had been hospitalised in a foreign country or in a Swiss hospital with high MRSA prevalence within the past 6 months indicating good adherence with that part of the guidelines (table 1).
The presence of wounds in 80% of MRSA-positive patients, whether diagnosed by screening or clinically indicated sample, might suggest extending microbiological MRSA admission screening to all patients with wounds or other skin conditions. However, due to the high frequency of such conditions in the general population, this would markedly increase the number of swabs taken and very likely augment the number needed to screen in our low prevalence setting, thus challenging cost effectiveness.
Data from MRSA patients newly diagnosed in the year after the one-year surveillance confirmed the notion that IVDU and tracheostoma/intubation do not play an important role as risk factors for MRSA carriage in our setting. Accordingly, in September 2010, we eliminated them from the algorithm depicted in figure 1, especially bearing in mind that simplification of the scheme might have a positive effect on compliance.
Following the phrase “Seek and you will find”, one could argue for routine universal admission screening. However, the cost-effectiveness of such a policy is unknown [19, 31, 32]. Thus, it appears to be more reasonable to invest available resources into improvement of hand hygiene compliance (48 to 66% in Pittet et al.) [33], which has been shown to simultaneously reduce the rate of MRSA transmission and the overall rate of healthcare-associated infections [33, 15].
In the sense of “keeping an eye on it”, screening high-risk patients for MRSA on hospital admission remains an acceptable compromise for MRSA surveillance in a low prevalence region, though this still leaves us with the question of who exactly are these individuals.
Adherence to the screening guidelines was not measured. Consequently, more admission screenings might have been indicated than eventually performed and we cannot prove that the 161 screened individuals were a fully representative sample of the admitted patients.
Due to the low prevalence setting and the restriction of MRSA admission screening to high risk patients, the number of positive screened patients was relatively small.
The generalisability of our findings is limited. In general, the diversity of settings (MRSA prevalence, compliance with standard precautions/hand hygiene) warrants individual guidelines and their appropriateness should be locally evaluated after introduction.
In the setting of low MRSA prevalence, hospital admission screening of patients at high risk for MRSA carriage detected only one of five newly diagnosed MRSA patients. Therefore, standard precautions, particularly hand hygiene, remain the most efficient measure to prevent MRSA transmission.
With 80%, the prevalence of wounds was high among MRSA positive patients. Most of them can be diagnosed by screening the nose, throat and wound.
1 Diekema DJ, Pfaller Ma, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M; SENTRY Participants Group. Survey of infections due to Staphylococcus species: frequency of occurence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin Amercia, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32(Suppl 2):S114–132.
2 Gould IM. The clinical significance of methicillin-resistant Staphylococcus aureus. J Hosp Infect. 2005;61(4):277–82.
3 Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003;36(1):53–9.
4 Bradley SF, Terpenning MS, Ramsey MA, Zarins LT, Jorgensen KA, Sottile WS, Schaberg DR, Kauffman CA. Methicillin-resistant Staphylococcus aureus: colonization and infection in a long-term care facility. Ann Intern Med. 1991;115(6):417–22.
5 Jernigan JA, Clemence MA, Stott GA, Titus MG, Alexander CH, Palumbo CM, Farr BM. Control of methicillin-resistant Staphylococcus aureus in a university hospital one decade later. Infect Control Hosp Epidemiol. 1995;16(12):686–96.
6 Marschall J, Mühleman K. Duration of methicillin-resistant Staphylococcus aureus carriage, according to risk factors for acquisition. Infect Control Hosp Epidemiol. 2006;27(11):1206–12.
7 Muder RR, Brennen C, Wagener MM, Vickers RM, Rihs JD, Handcock GA, et al. Methicillin-resistant Staphylococcus aureus: colonization and infection in a long-term care facility. Ann Intern Med. 1991;114(2):107–12.
8 Davis KA, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin Infect Dis. 2004;39(6):776–82.
9 EARSS (European Antimicrobial Resistance Surveillance System). Proportion of MRSA isolates in participating countries in 2008. Available at: http://www.rivm.nl/earss/database/
10 Swiss Centre for Antibiotic resistance. Antibiotic resistance data 2008: Oxacillin-resistance of Staphylococcus aureus. Available at: http://www.anresis.ch/de/index.html
11 Diekema DJ, Edmond MB. Look before you leap: active surveillance for multidrug-resistant organisms. Clin Infect Dis. 2007;44(8):1101–7.
12 Bootsma MC, Diekmann O, Bonten MJ. Controlling methicillin-resistant Staphylococcus aureus: Quantifying the effects of interventions and rapid diagnostic testing. PNAS. 2006;103(14):5620–5.
13 Wernitz MH, Swidsinski S, Weist K, Sohr D, Witte W, Franke KP, et al. Effectiveness of a hospital-wide selective screening programme for methicillin-resistant Staphylococcus aureus (MRSA) carriers at hospital admission to prevent hospital-acquired MRSA infections. Clin Microbiol Infect. 2005;11(6):457–65.
14 Peterson LR, Diekema DJ. To screen or not to screen for Methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2010;48(3):683–9.
15 Wenzel RP, Bearman G, Edmond MB. Screening for MRSA: a flawed hospital infection control intervention. Infect Control Hosp Epidemiol. 2008;29:1012–8.
16 Wertheim HF, Vos MC, Boelens HA, Voss A, Vandenbroucke-Grauls CM, Meester MH, et al. Low prevalence of methicillin-resistant Staphylococcus aureus (MRSA) at hospital admission in the Netherlands: the value of search and destroy and restrictive antibiotic use. J Hosp Infect. 2004;56(4):321–32.
17 Jones G, Sutton JK, Aplin S, Browning D. How many patients do you need to screen for methicillin-resistant Staphylococcus aureus to find a positive? Results from a screening programme in a UK teaching hospital. Clin Microbiol Infect. 2010;16(S2):S46.
18 Huang SS, Yokoe DS, Hinrichsen VL, Spurchise LS, Datta R, Miroshnik I, et al. Impact of routine intensive care unit surveillance cultures and resultant barrier precautions on hospital-wide methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis. 2006;43(8):971–8.
19 Harbarth S, Fankhauser C, Schrenzel J, Christenson J, Gervaz P, Bandiera-Clerc C, et al. Universal screening for methicillin-resistant Staphylococcus aureus at hospital admission and nosocomial infection in surgical patients. JAMA. 2008;299(10):1149–57.
20 Stelfox HT, Bates DW, Redelmeier DA. Safety of patients isolated for infection control. JAMA. 2003;290(14):1899–905.
21 Morgan DJ, Diekema DJ, Sepkowitz K, Perencevich EN. Adverse outcomes associated with contact precautions: a review of the literature. Am J Infect Control. 2009;37(2):85–93.
22 Cooper BS, Stone SP, Kibbler CC, Cookson BD, Roberts JA, Medley GF, et al. Systematic review of isolation policies in the hospital management of methicillin-resistant Staphylococcus aureus: a review of the literature with epidemiological and economic modelling. Health Technol Assess. 2003;7(39):1–194.
23 Mathews AA, Thomas M, Appalaraju B, Jayalakshmi J. Evaluation and comparison of tests to detect methicillin resistant S. aureus. Indian J Pathol Microbiol. 2010;53(1):79–82.
24 Brown DF, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, Wren MW on behalf of the Joint Working Party of the British Society for Antimicrobial Chemotherapy, Hospital Infection Society and Infection Control Nurses Association. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J Antimicrob Chemother. 2005;56(6):1000–8.
25 Fleisch F, Oechslin EC, Gujer AR, Ritzler E, Imhof A, Ruef C, Reinhart WH. Transregional spread of a single clone of methicillin-resistant Staphylococcus aureus between groups of drug users in Switzerland. Infection. 2005;33(4):273–7.
26 Edmond MB, Lyckholm L, Diekema DJ. Ethical implications of active surveillance cultures and contact precautions for controlling multidrug resistant organisms in the hospital setting. Public Health Ethics. 2008;1:235–45.
27 Sax H, Allegranzi B, Chraiti MN, Boyce J, Larson E, Pittet D. The World Health Organization hand hygiene observation method. Am J Infect Control. 2009;37(10):827–34.
28 Bühlmann M, Bögli-Stuber K, Droz S, Mühlemann K. Rapid screening for carriage of Methicillin-resistant Staphylococcus aureus by polymerase chain reaction and associated costs. J Clin Microbiol. 2008;46(7):2151–4.
29 Swiss-Noso 12/1995; Band 2, Nummer 4. Methizillin-resistenter S. aureus: Aktuelle Situation und Bedeutung. Available at: http://www.chuv.ch/swiss-noso/cd24a1.htm
30 Mertz D, Reno F, Periat N, Zimmerli M, Battegay M, Flückiger U, Widmer AF. Exclusive Staphylococcus aureus throat cariage. Arch Intern Med. 2009;169(2):172–8.
31 Robicsek A, Beaumont JL, Paule SM, Hacek DM, Thomson RB Jr, Kaul KL, et al. Universal surveillance for methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Ann Intern Med. 2008;148(6):409–18.
32 Weber SG, Huang SS, Oriola S, Huskins WC, Noskin GA, Harriman K, et al. Legislative mandates for use of active surveillance cultures to screen for methicillin-resistant Staphylococcus aureus and vancomycin-reistant enterococci: position statement from the Joint SHEA and APIC Task Force. Infect Control Hosp Epidemiol. 2007;28(3):249–60.
33 Pittet D, Hugonnet S, Harbarth S, et al. Effectiveness of a hospital-wide programme to improve compliance with hand hygiene. Infection Control Programme. Lancet. 2000;356:1307–12.
We thank all affiliated hospitals contributing data: Rorschach, Flawil, Altstätten, Grabs, Heiden, Herisau, Uznach, Walenstadt, Wattwil and Wil.
MRSA diagnostic was performed in the Institute of Clinical Microbiology and Immunology in St. Gallen.
Part of the data has previously been presented as an abstract/poster at the ECCMID 2010 in Vienna (P1539).
All three authors have seen and approved the manuscript and have significantly contributed to the work. The manuscript has not been published and is not being considered for publication elsewhere.
Funding / potential competing interests: No financial support and no other potential conflict of interest relevant to this article were reported.