miRNAs and rheumatoid arthritis – promising novel biomarkers
DOI: https://doi.org/10.4414/smw.2011.13175
I
Duroux-Richard, C
Jorgensen, F
Apparailly
Summary
Biomarkers are indicators of biological conditions that can be detected and measured in body fluids or tissues. Biomarkers can be detectable before the clinical onset of the disease, and are thus useful for prognosis; they can be measured at early stages of the disease and are useful for stratification and classification of the disease and patients; they can be monitored along the disease course and used as indicators of risk factors and pharmacological response to treatment. Ideally, biomarkers should be sensitive, specific, have high predictive power, and be easily accessible. Rheumatoid arthritis (RA) is the most frequent chronic inflammatory disorder, affecting millions of people worldwide and leading to joint damage and substantial morbidity. RA is a heterogeneous disorder with a fluctuating clinical course and unpredictable prognosis. And although a large panel of biologics is available to clinicians, the main challenge remains to treat patients as early as possible with the most personalised therapy. Today, the most challenging issue in RA is the identification of biomarkers for early disease diagnosis and for prediction of drug response. Among molecules that can fulfil this expectation, micro(mi)-RNAs certainly represent an option. The potential value of miRNAs as a novel class of biomarkers is well documented in cancer. Moreover, the presence and stability of miRNAs in body fluids provide fingerprints that can serve as molecular biomarkers for disease diagnosis and therapeutic outcome. As a growing body of evidences reveals abnormal expression of specific miRNAs in RA tissues, the use of a blood-based miRNA signature for optimal diagnosis and treatment becomes a realistic option.
Introduction
Rheumatoid arthritis (RA) is the most frequent inflammatory rheumatism. RA is a chronic, heterogeneous disease of uncertain aetiology that affects more than 2.9 million Europeans. This autoimmune disease is characterised by chronic inflammation within the joint tissue infiltrated by activated immune cells and by synovial hyperplasia, leading to cartilage and bone destruction after several years. The clinical course of RA fluctuates and the prognosis is unpredictable, 70% of patients with RA of recent onset show evidence of radiographic changes within 3 years [1]. RA is a progressive, debilitating disease with severe physical and economic consequences, work disability and often premature death [2]. A better understanding of the pathophysiology of RA has led to important innovative approaches to its treatment. Currently, although a large panel of biologics is available to clinicians, the main challenge remains to treat patients as early as possible with a therapy that will best fit their dominant disease subtype.
Current challenges for the treatment of rheumatoid arthritis
At present, 95% of newly diagnosed RA patients start with methotrexate monotherapy, which is ineffective in 66% of patients, and then are switched to (or given in association with) other disease-modifying anti-rheumatic drugs (DMARD) [3]. Treatment with conventional DMARDs is often associated with various adverse reactions due to unspecific and toxic properties [4]. When disease activity cannot be controlled, genetically engineered proteins targeting specific components of the immune system are used [5]. Such biologics, together with an improved timing and dosing of conventional therapy, have largely improved the outcome of established arthritis in many patients [6, 7], but not all, and still pose significant problems [8]. At present a 20% improvement by American College of Rheumatology criteria (ACR20 response rate) can be achieved in 60–70% of patients treated with anti-TNF biologics [9], but the majority of these responders still have some actively inflamed joints and complete remission is rare [10]. One major problem of the treatment with biologics is the need for ongoing systemic therapy that provides limited effects in many patients, with serious potential side effects including increased susceptibility to infections and uncontrolled fevers as well as cardiovascular disorders [11], and very high cost (EUR
15 000/patient/year), making these new therapies available to a restricted number of patients. Clinical and serological characteristics alone are insufficient to predict disease outcome. There are clear unmet medical needs in RA, and one of the most challenging issues is the identification of biomarkers for early disease diagnosis and therapeutic outcome. Among molecules that possess such potential, micro(mi)-RNAs certainly represent an interesting option to be explored.
Biogenesis of miRNAs
From nematodes to Drosophila or mammals, miRNAs are an abundant class of endogenous, short non-coding, single-stranded RNA molecules (19-23 nucleotides) that function as negative regulators of the expression of protein-encoding genes. Human miRNA genes are located in all chromosomes except Y chromosome and they are non randomly distributed in the human genome [12]. The majority of miRNA genes (70%) are found in the introns, in the sense orientation, and approximately 30% are located in intergenic regions. Approximately 50% of known human miRNAs are found in clusters and are transcribed by RNA polymerase II as mono- or poly-cistronic long primary transcripts, namely primary miRNA or pri-miRNA [13], ranging from approximately 200 nucleotides (nt) to several kilobases (kb) in length and folded into hairpin structures containing imperfectly base-paired stems (fig. 1). There are usually two or three genes per cluster and the largest cluster is composed of seven genes. Clustered miRNAs can be functionally related by targeting the same gene or different genes involved in the same metabolic pathway. In the nucleus, pri-miRNAs are cleaved by the DROSHA RNase III endonuclease into 70-100 nt-long hairpin with a monophosphate at the 5’ terminus and a 2-nt overhang with a hydroxyl group at the 3’ terminus called pre-miRNA [14]. The pre-miRNAs are transported to the cytoplasm by the Exportin-5 and its RAN-GTP cofactor [15]. The pre-miRNAs are further digested by Dicer, another RNase III endonuclease, along with its partner protein TRBP (trans-activator RNA binding protein) to produce the mature miRNA/miRNA* duplex of 20-22 nt [16]. Dicer cuts both strands at about two helical turns away from the base of the stem loop of the pre-miRNA. The duplex is unwound by a helicase and the miRNA “guide” strand, currently identified as the 5’ terminus that is energetically less stable, is then selected for incorporation into the RISC (RNA-induced silencing complex), while the “passenger” strand (miRNA*) is released and degraded [17]. Occasionally, both arms of the pre-miRNA hairpin give rise to mature miRNAs.
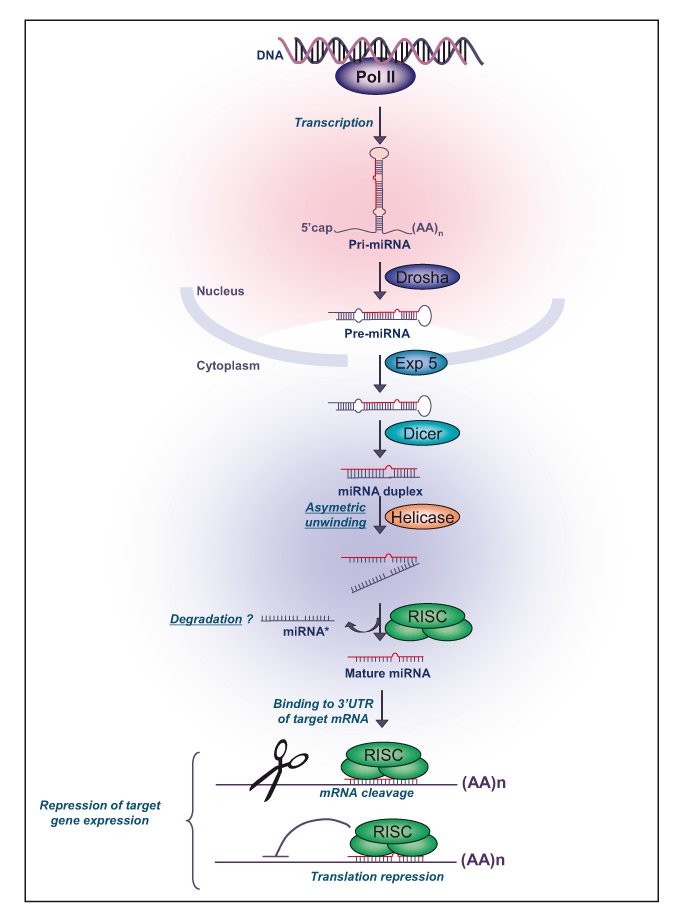
Figure 1: MicroRNA biogenesis.
Primary miRNA are transcribed by RNA polymerase II, processed by the RNase III DROSHA into 70–100 nt-long hairpin, and exported into the cytoplasm complexed with exportin-5. Once in the cytoplasm, the pre-miRNA is digested by Dicer to produce the mature miRNA duplex of 20–22 nt. The miRNA-RISC complex interacts with the complementary 3’- or 5’-untranslated region (UTR) of target messenger (m)RNAs and subsequently destabilizes them, or block their translation.
miRNA: microRNA; pri-miRNA: primary miRNA; pre-miRNA: precursor miRNA; RISC: RNA-induced silencing complex.Pol II polymerase II, Exp 5: Exportin 5
MicroRNA mode of action
Following cleavage by Dicer, the miRNA pathway is biochemically very similar to the RNA interference (RNAi) known as an RNA silencing pathway widespread from plants to animals. The actions of the miRNA guide strand are mediated by the catalytic component of the ribonucleoprotein (miRNP) RISC complex formed by a member of the Argonaute family (Ago) and a number of unknown accessory factors. RISC acts to downregulate the translation of target mRNA by mechanisms of
either mRNA cleavage or translational repression, depending on the perfect or incomplete complementary between the miRNA guide strand and its mRNA target. It is believed that most of the miRNAs typically mediate translational repression rather than mRNA cleavage. Among the Ago family proteins, only Ago2 is able to cleave the target by endonuclease activity [18]. All other Ago proteins mediate translational repression by imperfect complementary between the miRNA guide strand and the target mRNAs. In that case the expression of targeted genes is evidenced at the protein level without decreased mRNA levels. Alternatively, repressed mRNAs are relocated for storage into specific intracellular organelles called P-bodies, where miRNAs and associated proteins are co-localised [19].
In general, the complementary miRNA binding sites are located in the 3’ untranslated regions (UTRs) of the target mRNAs. Recent publications, however, show and predict that miRNAs might also interact with complementary sequences residing in the 5’UTR or the promoter and even the CDS regions of target genes [20–22]. Based on computational predictions, different miRNAs are believed to regulate the same targets, either on different sites or competing for the same location, leading to the concept of co-action or competition. For a miRNA to exert down regulation of gene expression, the most stringent requirement is a contiguous and perfect base pairing of the miRNA nucleotides 2–8, representing the “seed” region which nucleates the interaction, with the target mRNA sequence. The “seed” rule and its conservation among species are among the criteria used by computational miRNA target-gene prediction methods freely available on the internet [23]. They present limitations, however, showing that our actual knowledge of the mode of action of miRNAs is still incomplete and that many exceptions to the rules might exist.
Biological significance of miRNA-mediated gene regulation
There are at least 1000 predicted miRNAs in the human genome, representing ~0.0001% of the total genome, and predicted to regulate the expression of 10–60% of our protein-encoding genes, depending on the target prediction method used. This gene silencing pathway that regulates genetic programmes at the post-transcriptional level in a sequence-specific manner thus appears to play an important role in regulating gene expression and adds another layer of complexity to already known regulatory systems. By controlling the accumulation of the target protein(s) in cells, miRNAs have key functions in many physiological networks. They are involved in the regulation of almost every cellular process investigated so far, including differentiation, apoptosis, proliferation and metabolism [24]. Some miRNAs are widely expressed (miR15 and miR16) while others are expressed in a tissue-specific manner (miR122 and miR-142-3p) [25]. The expression of miRNAs is dynamically regulated during development, and although the function of most of the mammalian miRNAs has yet to be determined it appears that deregulation of this genetic regulatory network is associated with serious human disorders, including cancer. miRNA-encoding genes are commonly located within cancer-associated regions and a specific nomenclature has emerged to describe their implication in cancers, including oncomir, metastamir, apoptomir. It is therefore likely that miRNAs are also implicated in the pathogenesis of many human diseases, for which they might have promising biomedical potential [26].
miRNAs in rheumatoid arthritis
In 2007, the first indirect evidence was provided by a publication linking a key cellular component of the RNAi machinery (Argonaute 2) to RA. Indeed, autoantibodies detected in the serum of RA patients showed specificity for the Ago2 protein and other components of the P-bodies, a unique cytoplasmic structure involved in mRNA processing and RNAi [27]. From 2008, when three studies reported the up-regulation of specific miRNAs either in the circulation (miR-16, miR-132, miR-146a and miR-155, [28]) or within the inflamed joints (miR-146a and miR-155, [29, 30]), up to now the abnormal expression of 10 miRNAs has been documented in RA patients (table 1, [28–37]). Three of them are downregulated and six upregulated, with several convergent studies confirming the upregulation of miR-16, miR-146a, miR-223 and miR-155. With only one miRNA, studies have shown apparently conflicting results, miR-132 being highly expressed in PBMCs isolated from RA blood [28] but decreased in plasma samples [34] as compared with control donors. This apparent discrepancy is perhaps due to the fact that miRNAs expressed by blood cells are not necessarily found in plasma or serum. In fact, only a few of them are detectable in serum or plasma, and this will be the subject covered by the following paragraph. Although a role for miR-132 has been mainly described in neuronal morphogenesis, recent work suggests a regulatory role in innate antiviral immunity [38]. Lagos et al. showed that miR-132 mediates an antiinflammatory effect by inhibiting the expression of the p300 transcriptional co-activator, thus impairing the downstream expression of IFN-β, ISG15, IL-1β and IL-6.
The abnormal abundance of miR-16, miR-146a, miR-223 and miR-155 both systemically and within inflamed RA joints is not really surprising. Indeed, miR-16 and miR-223 are among the most abundant miRNAs in the blood, probably because of their higher expression levels by PBMCs, and miR-146a and miR-155 are shown upregulated in several immune-mediated inflammatory disorders. Indeed, miR-16 is ubiquitously expressed at high levels and several groups evidenced dysregulated expression of the miR-15/16 clusters in various lymphoid and myeloid malignancies. This cluster plays very
important roles in regulating cell proliferation and apoptosis by targeting cell cycle proteins and the antiapoptotic genes. Its high expression levels in RA blood [28, 34] might reflect an increase in global cellularity in blood, a systemic enrichment of specific haematopoietic lineages and/or a true deregulated expression of miR-16 in specific cell types. Interestingly, two groups have correlated a higher expression level of miR-16 in plasma [34] or PBMCs [28] from RA patients as compared with healthy donors, with DAS28, suggesting that miR-16 might be used as a biomarker of RA disease activity.
Serum miR-223 is already proposed as a potential biomarker for sepsis [39] and is now shown expressed in higher levels in RA samples than in OA tissues, and inversely correlated with tender joint count [34]. miR-223 is chiefly known for its role in innate immunity, mainly in granulopoiesis, and upregulation of miR-223 is documented in myeloid leukaemia [40]. However, miR-223 is enriched in resting CD4+ T cells as compared to activated CD4+ T cells, and Fulci and coworkers showed that miR-223 is the only miRNA that is markedly upregulated in peripheral naive CD4+ T-lymphocytes from RA patients compared with healthy donors [32].
Finally, miR-146a and miR-155 are the most frequently reported miRNAs deregulated in RA samples including blood, plasma [34], synovial fluid, PBMCs [28], CD4+ T cells [33, 36] isolated from the blood or the synovial fluid, or RA-FLS [29, 30]. Increased miR-146a expression levels are correlated with active disease in RA patients [28]. They are both involved in the development of innate and adaptive immune cells, and numerous studies report their upregulation in inflammatory conditions [41, 42]. Both miR-146a and miR-155 are induced in lymphoid and myeloid cells in response to a variety of microbial components and pro-inflammatory cytokines, where they are considered to play a role in fine-tuning immune responses through negative feedback loops on genes induced by inflammatory cascades [43]. It is thus not really surprising that first studies on miRNAs in RA investigated these two specific miRNAs.
|
Table 1: microRNAs deregulated in rheumatoid arthritic tissues. |
|
|
miR16
|
miR124a
|
miR132
|
miR146a
|
miR155
|
miR203
|
miR223
|
miR346
|
miR363
|
miR498
|
| Joint |
ST |
+ |
|
+ |
|
+ |
|
+ |
|
|
|
| FLS |
|
– |
|
+ |
+ |
+ |
|
+ |
|
|
| SF |
+ |
|
+ |
+ |
+ |
|
|
|
|
|
| CD4+ |
|
|
|
+ |
|
|
|
|
- |
- |
|
Ref. |
(34) |
(35) |
(34) |
(29,30,34) |
(30,34) |
(37) |
(32,34) |
(31) |
(33) |
(33) |
| Blood |
PBMC |
+ |
|
+ |
+ |
+ |
|
|
|
+ |
|
| Serum Plasma |
+ |
|
– |
+ |
+ |
|
+ |
|
|
|
| CD4+ |
|
|
|
+ |
|
|
+ |
|
– |
- |
|
Ref. |
(28,34) |
|
(28,34) |
(28,33,34,36) |
(28,34) |
|
(32,34) |
|
(33) |
(33) |
| Abbreviations: ST, synovial tissue; FLS, fibroblast-like synoviocytes; SF, synovial fluid; CD4+, T cell lymphocytes positive for the CD4 phenotypic marker; and PBMC, peripheral blood mononuclear cells. (+) increased or (-) decreased expression levels |
Detection of miRNAs in body fluids
Since 2008 several studies have evidenced the possibility of detecting miRNAs in body fluids including serum, plasma, urine, saliva, tears, amniotic and placental fluids, thus opening up major opportunities for a novel type of diagnostic molecules [44–46]. Since RA is a systemic chronic inflammatory disorder for which peripheral blood gene expression signature has been reported and molecular biomarkers are of great interest, identification of miRNA-based signatures is indeed a major issue. Most of the studies supporting the clinical utility of miRNAs as biomarkers in body fluids or diseased tissues have been conducted in cancer. High concentrations of cell-free miRNAs originating from the primary tumour have been found in the plasma of cancer patients, and several lines of evidence indicate that circulating miRNAs represent a promising source of cancer biomarkers [47]. Indeed, correlations between miRNA expression levels and the development of malignancies, disease severity and aggressiveness, metastatic potential, therapeutic response and survival are reported in various cancer types. Interestingly, tumour-associated miRNomes appear highly tissue-specific.
The detection of miRNAs in serum was quite unexpected as RNA molecules are unstable in the circulation. Studies [44, 46, 48] showed that miRNAs exhibit high stability in the serum and plasma as they circulate within membrane vesicles such as exosomes or microparticles which protect them from endogenous RNase activity [49]. These microvesicles are resistant to drastic conditions and express tissue-specific markers. Although it is easily accessible and of great interest for new biomarker discovery, very few studies report the optimisation of extraction and detection of miRNAs in plasma or serum [45, 46]. Several technical challenges in miRNA extraction, detection and quantification in serum or plasma are still underinvestigated. More-over, all the miRNAs detected in total blood samples are not found in serum or plasma, and/or the low concentration of most of the miRNAs in serum or plasma precludes their detection, thus limiting the panel of miRNAs for solid profiling. Indeed, it is currently estimated that 20 miRNAs are needed for solid definition of biomarker signatures. For all these reasons, and mainly technical reasons, whole blood profiling is to be definitively the main focus in future studies on biomarker discovery, as opposed to plasma- or serum-based signatures. Although it seems to be very close, many more studies are needed before the use of miRNAs can fulfil criteria for their use as reliable tools in diagnostic and prognostic settings.
miRNAs as biomarkers for RA management
Currently, autoantibodies, markers of cartilage degradation and inflammation, genetic and transcriptomic markers have been identified in RA. However, due to the lack of reliable markers for classification and prediction of disease outcome in RA, the search for additional biomarkers is highly needful. Although the potential value of miRNAs as molecular biomarkers for diagnosis, prognosis of disease outcome, and prediction of therapeutic response is widely documented in cancer, it is still largely unexplored in RA. The identification of abnormal miRNA expression in the circulation or inflamed joints of RA patients is still in its infancy and needs to be developed. Many more studies will be required to establish a miRNA-based signature in the circulation of RA patients that could be used by clinicians as biomarkers, not only in RA but also in other rheumatic disorders including OA [50]. Until now there has been only one publication in which the concentrations of 5 miRNAs in RA patients’ body fluids are measured, bringing the first proof for the use of miRNA biomarker potential in RA [34]. However, this study shows that only miR-132 plasma concentrations were significantly lower in RA than in healthy donors, but certainly not useful as diagnostic biomarkers since not correlated with DAS28 (28-joint Disease Activity Score) and not disease-specific since also significantly lower in OA plasma than in healthy donors. More urgently than diagnosis, biomarkers predicting drug efficacy are definitely required for optimal management of RA patients.
Correspondence to:
Florence Apparailly PhD
Inserm U844
CHU Saint Eloi
bâtiment INM
80 rue Augustin Fliche
F-34295 Montpellier cedex 5
France
florence.apparailly@inserm.fr
References
1 van der Heijde DM, van Leeuwen MA, van Riel PL, Koster AM, van ’t Hof MA, van Rijswijk MH, et al. Biannual radiographic assessments of hands and feet in a three-year prospective followup of patients with early rheumatoid arthritis. Arthritis Rheum. 1992;35(1):26–34.
2 Pincus T. The underestimated long term medical and economic consequences of rheumatoid arthritis. Drugs. 1995;50(Suppl 1):1–14.
3 van der Kooij SM, de Vries-Bouwstra JK, Goekoop-Ruiterman YP, van Zeben D, Kerstens PJ, Gerards AH, et al. Limited efficacy of conventional DMARDs after initial methotrexate failure in patients with recent onset rheumatoid arthritis treated according to the disease activity score. Ann Rheum Dis. 2007;66(10):1356–62.
4 Kirwan J. Adverse effects of low-dose glucocorticoids and DMARD therapy in patients with RA – a complex relationship? Nat Clin Pract Rheumatol. 2008;4(11):568–9.
5 Feldmann M, Williams RO, Paleolog E. What have we learnt from targeted anti-TNF therapy? Ann Rheum Dis. 2011;69(Suppl 1):i97–9.
6 McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 2007;7(6):429–42.
7 Genovese MC, Bathon JM, Fleischmann RM, Moreland LW, Martin RW, Whitmore JB, et al. Longterm safety, efficacy, and radiographic outcome with etanercept treatment in patients with early rheumatoid arthritis. J Rheumatol. 2005;32(7):1232–42.
8 Bongartz T, Sutton AJ, Sweeting MJ, Buchan I, Matteson EL, Montori V. Anti-TNF antibody therapy in rheumatoid arthritis and the risk of serious infections and malignancies: systematic review and meta-analysis of rare harmful effects in randomized controlled trials. Jama. 2006;295(19):2275–85.
9 Genevay S, Finckh A, Ciurea A, Chamot AM, Kyburz D, Gabay C. Tolerance and effectiveness of anti-tumor necrosis factor alpha therapies in elderly patients with rheumatoid arthritis: a population-based cohort study. Arthritis Rheum. 2007;57(4):679–85.
10 Simard JF, Arkema EV, Sundstrom A, Geborek P, Saxne T, Baecklund E, et al. Ten years with biologics: to whom do data on effectiveness and safety apply? Rheumatology. (Oxford) 2011;50(1):204–13.
11 Gabriel SE. Tumor necrosis factor inhibition: a part of the solution or a part of the problem of heart failure in rheumatoid arthritis? Arthritis Rheum. 2008;58(3):637–40.
12 Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8.
13 Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21(17):4663–70.
14 Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9.
15 Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6.
16 Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132(21):4645–52.
17 Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–6.
18 Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–86.
19 Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15(23):2149–55.
20 Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5‘UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–71.
21 Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci. U S A 2007;104(23):9667–72.
22 Zhou X, Duan X, Qian J, Li F. Abundant conserved microRNA target sites in the 5‘-untranslated region and coding sequence. Genetica. 2009;137(2):159–64.
23 Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34.
24 Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5.
25 Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166.
26 Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15(12):2070–9.
27 Bhanji RA, Eystathioy T, Chan EK, Bloch DB, Fritzler MJ. Clinical and serological features of patients with autoantibodies to GW/P bodies. Clin Immunol. 2007;125(3):247–56.
28 Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10(4):R101.
29 Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–92.
30 Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–9.
31 Alsaleh G, Suffert G, Semaan N, Juncker T, Frenzel L, Gottenberg JE, et al. Bruton’s tyrosine kinase is involved in miR-346-related regulation of IL-18 release by lipopolysaccharide-activated rheumatoid fibroblast-like synoviocytes. J Immunol. 2009;182(8):5088–97.
32 Fulci V, Scappucci G, Sebastiani GD, Giannitti C, Franceschini D, Meloni F, et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol. 2010;71(2):206–11.
33 Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang Y, et al. Altered micro- RNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. 2010;12(3):R81.
34 Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12(3):R86.
35 Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1294–304.
36 Niimoto T, Nakasa T, Ishikawa M, Okuhara A, Izumi B, Deie M, et al. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet Disord. 2010;11:209.
37 Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, et al. Altered expression of miR-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63(2):373–81.
38 Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12(5):513-9.
39 Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, et al. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394(1):184–8.
40 Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115(9):1768–78.
41 Carissimi C, Fulci V, Macino G. MicroRNAs: Novel regulators of immunity. Autoimmun Rev 2009;8(6):520–4.
42 Duroux-Richard I, Presumey J, Courties G, Gay S, Gordeladze JO, Jorgensen C, et al. microRNAs as new player in rheumatoid arthritis. Join Bone Spine. 2011;78(1):17–22.
43 Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci. U S A 2006;103(33):12481–6.
44 Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006.
45 Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3(9):e3148.
46 Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. U S A 2008;105(30):10513–8.
47 White MA, Fatoohi E, Metias M, Jung K, Stephan C, Yousef GM. Metastamirs: a stepping stone towards improved cancer management. Nature Reviews Clinical Oncology. 2010;173.
48 Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390(1):1–4.
49 Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3(11):e3694.
50 Oliviero F, Ramonda R, Punzi L. New horizons in osteoarthritis. Swiss Med Wkly. 2010;140:w13098.
