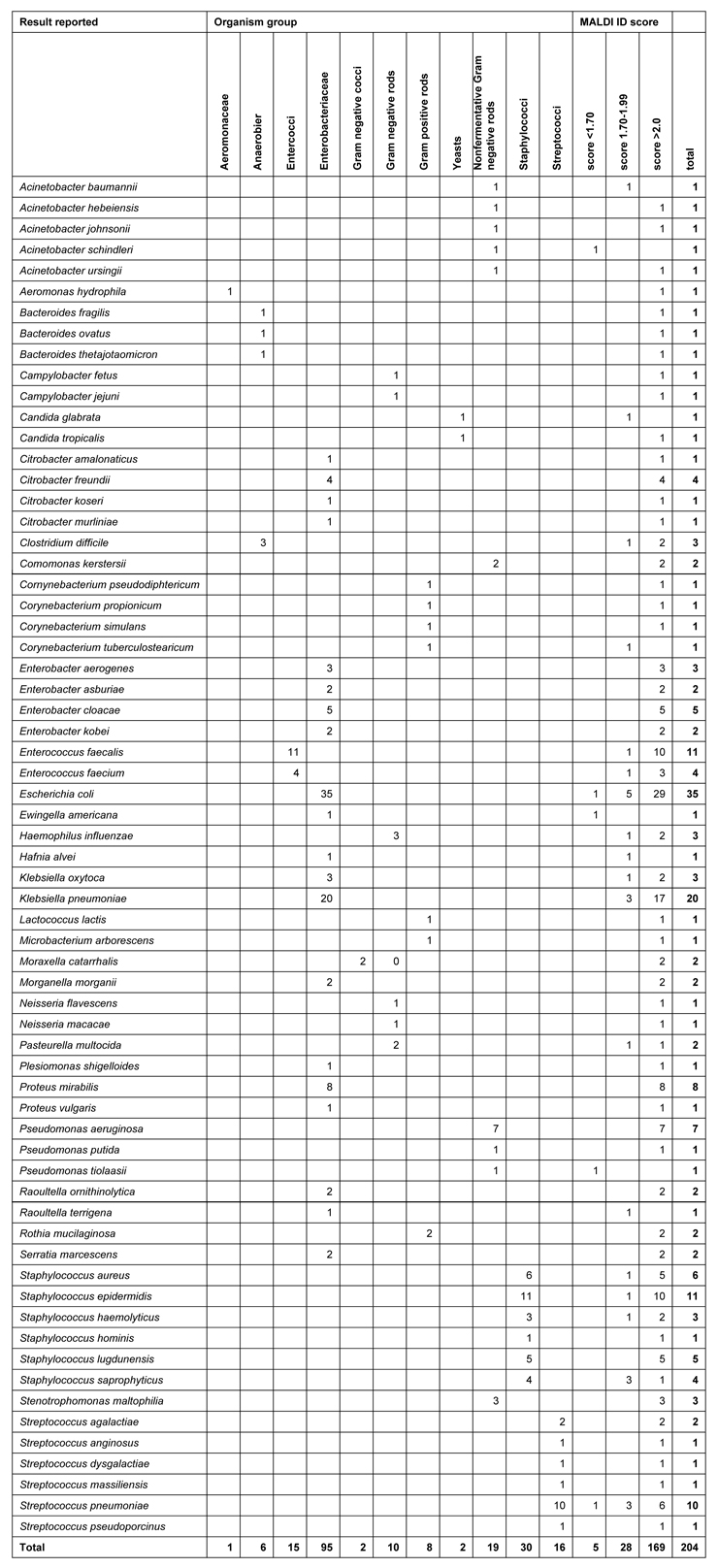
Table 1
Clinical isolate characterisation listed by organism group and MALDI identification score
DOI: https://doi.org/10.4414/smw.2010.13095
In addition to morphological, biochemical microbiological testing analysis along with molecular identification at the DNA and mRNA levels, the matrix assisted laser induced desorption ionisation (MALDI), connected to the time of flight (TOF) channel, is now becoming a third diagnostic pillar with strong discriminating power [1–4]. Thanks to the possibility to ionise and introduce non-volatile molecules into the mass spectrometer, this technique allows for classification and identification of micro-organisms through proteomic fingerprinting and comparison of the spectrometric peak pattern with the MALDI biotype data base which uses the BLAST (Basic Local Alignment Search Tool) procedure; the results are expressed using standard scores based on algorithms for specific identification of the biomarker proteins (SIBP).
A major thrust to use MALDI comes from its time saving benefit: the 24–48 h needed with culture-based methodologies is cut down to a few minutes. At our institution, a MALDI TOF analyser was introduced into the workflow as early as January 2009. At that time, only few studies were available which compared bacterial identification obtained on MALDI TOF with those resulting from conventional procedures; In light of this very new time-saving diagnostic tool, many health authorities have yet to approve this technology. At this early stage of MALDI TOF introduction into daily practice, comparison of microbes identified through this expedite procedure to conventional procedures seems mandatory; this paper reports the results of a comparative analysis on 204 clinical specimens for bacteriological analysis provided by MALDI TOF and by conventional BactAlert (API) and Vitek 2 biotype system.
The origin of 204 clinical samples was the following: 66 samples were urine specimens, 17 positive blood cultures, 27 genital specimens, 27 wound swabs, 23 faecal specimens and the remainder were 6 sputum and various other samples taken under precaution to avoid commensal contamination.
All isolates from the different origins were recovered after aerobic, microaerophilic and anaerobic incubation of clinical specimens. As a bacterial culture medium, citrated sheep blood agar was used with the CPDA-1 adenine-enriched formula. Growth of colonies was ascertained after overnight incubation by visual inspection. Blood cultures were applied at the patients bed side to BacT/ALERT® FN culture flasks and placed at our institution for 7 days in the automated BacT/ALERT® system (bioMérieux, Inc, Durham, NC; USA).
The conventional identification of microbes comprised two biomerieux® products:
For VITEK® 2 ID testing the ID-GP and GN cards were used. Evaluation was done by colorimetric analysis by the apparatus relating to its data base provided by the supplier.
A sufficient number of colonies was suspended in sterile saline (0.45%) and the McFarland turbidity was adjusted to .45%.
The API20® strips were applied where indicated and API tests were performed according to the manufacturer’s instructions. The metabolic results were read using an ap 20E (biomérieux)(ref no 20 100/20 160) and api20NE.
With 6 cultivated germs, a 16S rDNA fragment (800 bp) was sequenced. For this purpose, samples were sent to the Institute for Medical Diagnosis (www.imdlab.ch Zurich, Switzerland) where DNA was extracted, 16S ribosomal RNA genes amplified and 16S rRNA gene clone libraries constructed and compared between groups. An isolate was correctly identified when its almost full-length 16S rRNA gene sequence yielded >98.7% sequence similarity with the closest bacterial species sequence in the GenBank.
The three procedures reveal their results as a printout of the identified germ corroborated by a concurrence score. These are calculated by the intrinsic BLAST software of analysers which compare the data of the analyte with the data bank.
In parallel to inoculation into both, the Vitek identification cards and API ANA strips, one single colony was directly deposited on a MALDI-TOF MTP 384 target plate (Bruker Daltonik GmbH). The preparations were overlaid with 1 µl of matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, and 2.5% tri-fluoracetic-acid). The matrix-sample was crystallised by air-drying at 20–24 °C for 5 min. Mass spectrometry, data acquisition and spectra standard pattern matching as described elsewhere in detail [1]. As for validation of the results, we used score values proposed by the manufacturer of the apparatus: a score of >1.9 indicated species identification, a score of 1.7–1.9 indicated genus identification and a score of <1.7 indicated no identification.
In order to explore potential intrinsic variations of results obtained on our MALDI TOF system, we first assessed its stability in the release of bacterial identification data. Three types of procedures were chosen.

Table 1
Clinical isolate characterisation listed by organism group and MALDI identification score
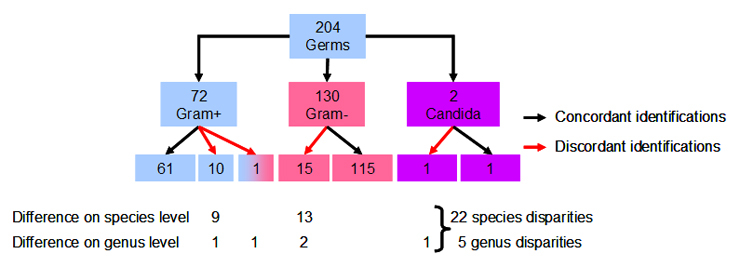
Figure 1
I. culture dependency. Cultures of Escherichia coli were grown on the two genus-specific differential media, one lactose-enriched and the other the gram-negative rod selective MacConkey agar plates. In 7 out of 7 experiments, the classification results revealed significant scores for Escherichia coli. Previously identified aerobic, gram-negative Acinetobacter ursingii and johnsoniicultures in lactose-monohydrate-enriched medium were applied to 2 and 4 sample positions on the metal MSP 96 ground plate of MALDI Bruker and 2 out of 2 results revealed the ursingii and 4 out of 4 revealed the johnsonii species of this proteobacterium with score values in 5 out of 5 identifications >2.0.
II. reproducibility assessment. The following clinical case showed significant reproducibility. Ten blood cultures growing Staphylococcus lugdunensiswere taken at 10 different time points and at different vascular catheter access sites of an 87 year old female suffering from bacterial sepsis in the course of surgery on her heart valves. The 10 different samples identified on the MALDI Bruker model revealed 10 times Staphylococcus lugdunensiswith a mean concurrence score value of 2.272 ± 0.134 (x ± 2 SD).
III. score/rank consistency. We analysed a sample of the fungal eukaryote Candida glabrata, previously identified using conventional procedures. The MALDI Biotyper revealed matched patterns for score values ranging from 1.659 to 1.396 in the first 4 rank places, a consistency that refutes the presence of another germ than Candida glabrata.
In order to assess identity/disparity of the results obtained with MALDI-TOF to conventional identification, we analysed the 204 clinical samples with both procedures. Of the 204 isolates identified, 72 were gram-positive and 130 were gram-negative strains and 2 were yeasts (candida). The correct MALDI identification, compared to conventional identifications was 86.8%. In 27 samples (31.2%), a disparity of the species and/or genus name became apparent: it appeared in 16 gram-negative (12.3%) and in 11 gram-positive germs (15.3%, n.s.). The characterisation of the clinical isolates is listed in the order of organism group on table 1.
The disparities in genus and species-assignments by MALDI TOF and conventional procedures (fig. 1) are as follows: in 14 of 27 discrepancies the disparity is of minor importance for patient and clinician, since the different identifications by MALDI TOF and conventional procedure are minor and comprise germs causing similar clinical pictures and exhibit the same antibiotic sensitivity pattern. The decision which result – conventional identification or MALDI TOF – is to be released to the clinician, is made on a case by case basis. In 13/27 disparities, the differential identities are major and clinically significant (table 2). Among them, 6 samples showed the same disagreement with Streptococcus pneumoniae (MALDI) and Streptococcus mitis/oralis (conventional identification) constellation. Among gram-negative samples, most instances exhibited a difference on the species level only, e.g. Enterobacter cloacae versus Enterobacter kobei.Twenty-two discordant results were seen on the species level. However, in 5 instances, the differences related to genus disparities: Enterobacter/Raoultella, Streptococcus/Gemella, Pseudomonas/Burkholderi, Microbacter/Sphingomonasand Candida/Cryptococcus. The most pertinent difference was Microbacterium arborescens (+) (MALDI) and Sphingomonas paucimobilis (–) (conventional identification).
With 6 clinical samples selected for their disparity between MALDI and conventional testing, the 16S rRNA-Gene PCR was performed on 880 base pairs and compared to results obtained with MALDI TOF and conventional identification.
In 2 out of these 6 isolates, the MALDI TOF results were confirmed. In another 2 samples a clinically irrelevant minor base-pair difference between the species alignements was seen, whereas in one sperm and one urinary sample the putative discrepancy between Streptococcus mitisand pneumoniae assignment appeared again (see above) (table 3).
The clinical advantage of accelerating microbial diagnosis to speed up specific antibiotic and supportive treatment will continue to encourage the introduction of faster, inexpensive and accurate methods for identifying bacterial isolates. Proteomics-based MALDI TOF for bacterial identifications are basically different from current routine practice based on phenotypic tests, culture and growth characteristics as well as biochemical patterns based on metabolomics. In 2009, the MALDI Biotyper obtained an IVD-CE mark with a Declaration of Conformity in accordance with the European Union in vitro Directive 98/79/ES. At the time of this report, the biotyper is not certified by Swiss Health Authorities for use in routine diagnosis which prompted us, despite its acceptance in the EU, to focus on reproducibility and comparability studies.
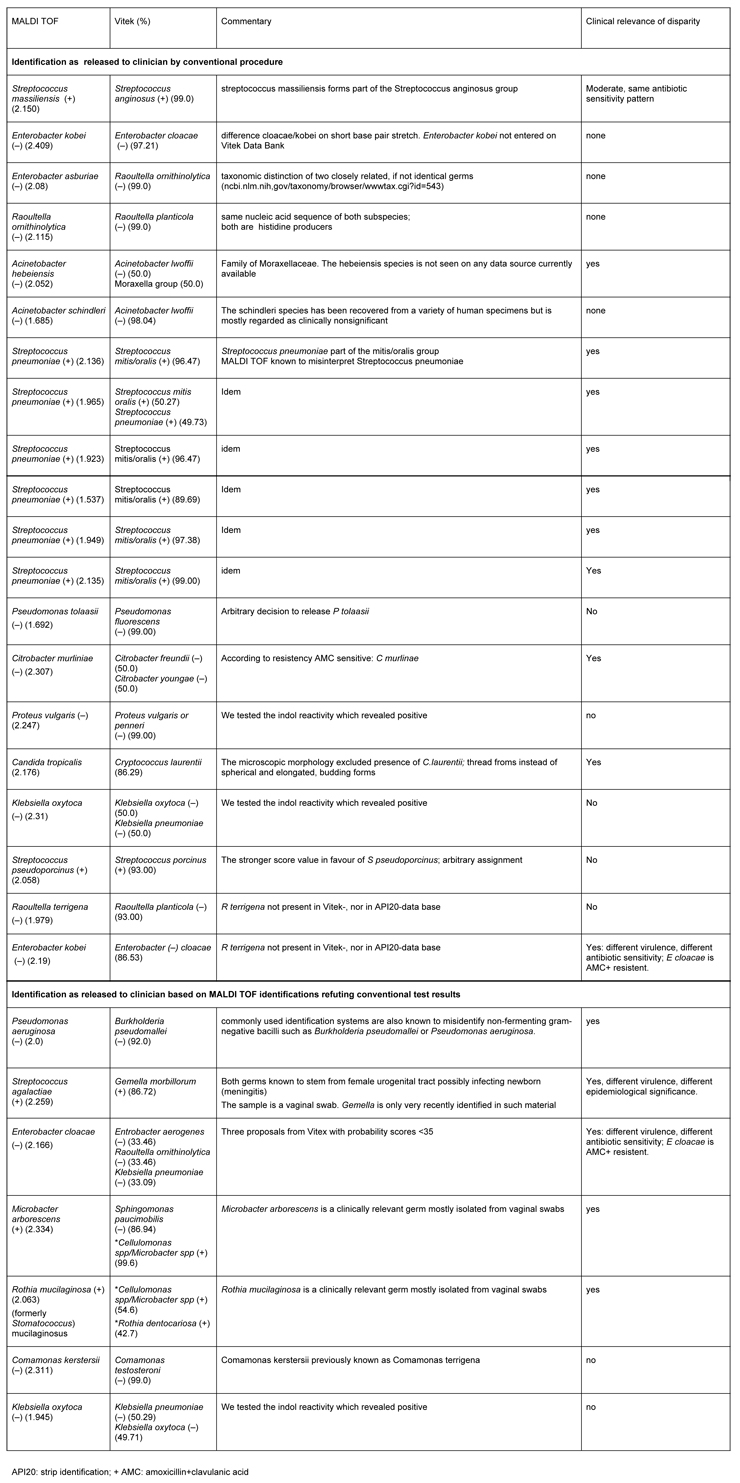
Table 2
Discordant results etween MALDI and Conventional Identification for 27 clinical isolates.
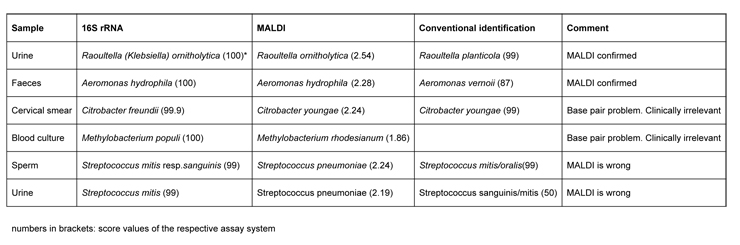
Table 3
Comparative analysis of bacterial identification including MALDI, conventional identification and 16S rRNA sequencing
Earlier comparative studies found similar correct-identification-rates, i.e., in agreement with conventional procedures; one study found similar comparisons to those found here [1] (84.1%) and another study [2] found 99.1% and a third study [3] found 93% versus 86.8% here. In the former study, the improved correlation is probably due to attributing 16S rDNA gene sequencing the value of a “gold standard”: the researchers thus were considering the molecular biological procedure as an axiomatic procedure to resolve the discordant findings. It is well known that different sequences within a short base pair stretch may question species alignment based on metabolomic identification and the confrontation of gene sequencing and MALDI TOF are a topic of recent studies [5]. One might recognise 16S rDNA sequencing as a gold standard bearing in mind that other standard identification methods provide essential information for the clinical relevance of a given germ. At least 16S rDNA genotyping does not form part of daily routine in most microbiology routine laboratories, whereas MALDI TOF is on its way to become so.
Although performed on a restricted number of clinical samples (204), our study focuses on the major clinically important germs currently encountered. Similar validations were performed by other study groups [2–4]. Most cases of misidentification are due to disparities at the species level and some are due to improper/incomplete database entries such as Raoultella planticola, an example observed, at the time of writing. With respect to clinical decision making, our observations direct special attention to the genus streptococcuson its species level.
Phylogenetic analysis based on 16S rRNA sequences of strain types of the more than 50 Streptococcusspecies revealed a clustering pattern with the mitis group standing apart: this group contains one of the leading pathogens in clinical pathology, Streptococcus pneumoniae along with 11 species that are prototype commensals of the upper respiratory tract; the problems of identification in clinical microbiology laboratories between the Streptococcus pneumoniae being strikingly similar to the three commensal species Streptococcus mitis, Streptococcus oralisand Streptococcus infantis. The distinction between Streptococcus pneumoniaeand Streptococcus mitis is difficult by definition, since the two species are genotypically close, unless a bile solubility test is used for differential diagnosis: pneumoccci dissolve, streptococci resist.
We cannot formally exclude transfer-related discordant results when the technician deposits culture aliquots to the target plate, although our working protocols implement written double-checks.
Recently published results from polyphasic phylogenetic analysis of genetic cluster studies disclose some of the pathways of the evolution crossed by pathogenic and commensal streptococci [6]. A recent study of 212 positive blood cultures representing 32 genera and 60 species failed to distinguish the mitisfrom the pneumoniaespecies of Streptococci [4] and MALDI TOF is well known to fail in producing significant matches of Streptococcus pneumoniaewith genome databases (http://hdl.handle.net/1853/5047) at least at present [4]. When 16S sRNA gene analysis was taken as a diagnostic starting point, none of the isolated germs revealed identical designations. It thus appears that a taxonomical difference prevails over the suspicion, that there was a true difference in germ identity even more so as in each of the 6 instances the genus specificity was identical across the board to genus identity. We cannot exclude, that a particular culture allowed for growth of two closely related germs, the one identified by 16S sRNA genotyping, the other by either one of the other two procedures.
The disparities seen on the species level may be of minor significance as regards the antibiotic resistance pattern, hence the importance of the clinical context. It is well known, that microbes of the same genus share their strategy in developing resistance using ribosome-derived nucleic acid sequences but subtle differences in individual germs may determine the extent of their virulence. Thus, virulent-phase cultures of Bordetella pertussis,but not other Bordetella species specifically recruit the complement system C1 inhibitor from human serum thus acquiring resistance [7]. In addition, Staphylcoccus aureussheds a 50 kDa component from its surface which increases factor I cleavage of C3b thereby compromising host resistance [8].
With respect to table 2a which lists the results released on the basis of conventional identification, our study supports the view that proteomics-dependent MALDI TOF should not completely outdate traditional metabolomic-dependent identification algorithms. These will be further needed in clinical microbiology, despite their shortcomings: time-consuming metabolic reactions, including resistance to antibiotic profiles, morphological and observer-dependent judgments. The future will confirm the economic impact of such widespread use of MALDI TOF. A recent report supports replacement of labour-intensive procedures by MALDI TOF [9]. Other endeavours adding value are under way: thus, broad-range PCR and microarray-based platforms designed to identify bacterial pathogens in positive blood cultures have proven sensitive, specific and almost as fast as MALDI, since they are based on amplification and detection of gyr B, par E, and mecA genes of the 50 most common bacterial species [10]. While such an approach might be integrated into everyday laboratory workflow in primary-care, its usefulness must still be proven for general care microbial diagnosis for which MALDI TOF seems more appropriate. The time saving approach now provided by the MALDI based protein fingerprinting of bacterial identity raises hope that daily rounds of treating physicians will further improve based on progress in the microbiology laboratory.
Mr G. Brighouse, MTA, Department of Pathology, University of Geneva, did linguistic editing.
1 Seng P, Drancourt M, Gouriet F, La Scola B, Forunier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–51.
2 Cherkaoui A, Hibbs, J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010;48(4):1169–75.
3 Bizzini A, Durussel C, Bille J, Greub G, Prod’hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48(5):1549–54.
4 Stevenson LG, Drake SK, Murray PR. Rapid identification of bacteria in positive blood culture broths by MALDI TOF mass spectrometry. J Clin Microbiol. 2010;48(2):444–7.
5 Mellmann A, Cloud J, Maier T, Keckovoet U, Ramminger I, Iwen P, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008;46(6):1946–54.
6 Kilian M, Polsen K, Blomqvist T, Havarstein LS, Bek-Thomsen M, Tettelin H, et al. Evolution of streptococcus pneumoniae and its close commensal relatives PloS On e 2008;3(7):e2683.
7 Marr N, Luu RA, Fernandez RC. Bordetella pertussis binds human C1 esterase inhibitor during the virulent phase, to evade complement-mediated killing. J Infect Dis. 2007;195(4):585–8.
8 Hair PS, Echague CG, Sholl AM, Watkins JA, Geoghegan JA, Foster TJ, et al. Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of staphylococcus aureus, and decreases complement-mediated phagocytosis. Infect Immun. 2010;78(4):1717–27.
9 Spanu T, De Carolis E, Fiori B, Sanguinetti M, D’Inzeo T, Fadda G, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to rpoB gene sequencing for species identification of bloodstream infection staphylococcal isolates. Clin Microbiol Infect 2010; Feb 2 (Epub ahead of print).
10 Tissari P, Zumla A, Tarkka E, Mero S, Savolainen L, Vaara M, et al. Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study. Lancet. 2010;375(9710):224–30.
Dr. Han was funded by grants from the Salvation Army.